Report on Omicron Spike mutations on epitopes and immunological/epidemiological/kinetics effects from literature
Authors: Anna Bernasconi*, Pietro Pinoli*, Ruba Al Khalaf, Tommaso Alfonsi, Arif Canakoglu, Luca Cilibrasi, and Stefano Ceri
Affiliation: Department of Electronics, Information, and Bioengineering, Politecnico di Milano, Milan, Italy
Notes: * The first two authors contributed equally to the work
The B.1.1.529 variant has entered abruptly the landscape of SARS-CoV-2 variants at the end of November 2021; it was reported to WHO on November 24th, named Omicron and considered a Variant of Concern, following the advice of the Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE). We report here the collection of our findings, following a long-standing effort of our group dedicated to SARS-CoV-2 sequences (involving data integration pipelines, search and knowledge management systems, see http://www.bioinformatics.deib.polimi.it/geco/?virus.
We collected Omicron Spike mutations of interest from ECDC (https://www.ecdc.europa.eu/en/covid-19/variants-concern) and compared them to CoVariants ones (https://covariants.org/variants/21K.Omicron). The provided lists only differ for the representation of the following deletions/insertions:
- G142D, Δ143-145 (ECDC) becomes G142-, V143-, Y144-, Y145D (CoVariants);
- Δ211-212, ins214EPE (ECDC) becomes N211- (CoVariants).
We then collected the Spike mutations of the three other currently designated Variants of Concern (Beta, Gamma, and Delta) from CoVariants, which provides a richer characterization (see https://covariants.org/variants/20H.Beta.V2, https://covariants.org/variants/20J.Gamma.V3, and https://covariants.org/variants/21A.Delta). We finally counted 37 amino acid changes for Omicron, 10 for Beta, 12 for Gamma, and 9 for Delta.
We then collected all linear SARS-CoV-2 B cell and T cell epitopes on the Spike protein, for human host species, curated by the Immune Epitope Database and Analysis Resource [A], available from the CSV Metric Exports (https://www.iedb.org/database_export_v3.php). Observing epitope variability has applications in disease monitoring, diagnostic settings, and drug design. A mutation occurring on the specific epitope range may affect the recognition of the epitope. The number of mutations on epitopes for a specific population of sequences can be computed using our EpiSurf tool [B].
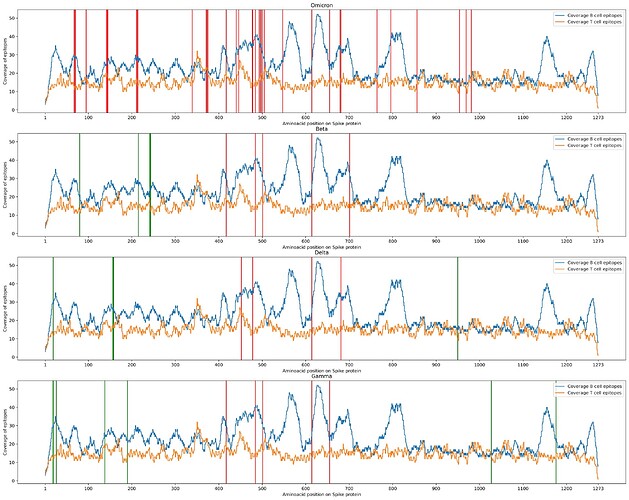
The figure below results from the comparison of the four Variants of Concern. For each of them, we computed a step-plot where for each position on the Spike protein (x-axis) we count (on the y-axis) how many SARS-CoV-2 linear epitopes are found in IEDB overlapping that position. The blue plot counts B cell epitopes, and the orange plot counts T cell epitopes. Along the protein, we plot mutations as vertical lines, red (if they were reported in the ECDC list) or green (if they were reported only in the CoVariants list). The plot highlights the difference of the 4 VoCs with respect to the number of salient Spike mutations; moreover, it shows that several Omicron mutations occur in Spike regions with a high concentration of B cell epitopes (i.e., 67-70, 142-145, 211-214, 477-505, 796).
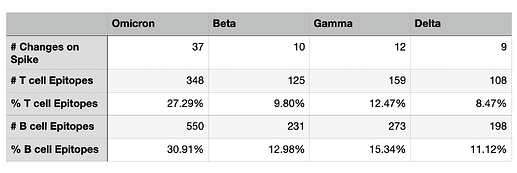
The table below shows quantitative statistics on the overall affected epitopes. Omicron mutations impact 348 IEDB T cell epitopes (27.29% of the total) and 550 IEDB B cell epitopes (30.91% of the total). Percentages of other Variants of Concern are significantly lower.
We conducted an extensive analysis of SARS-CoV-2 published literature and collected trustworthy references from COG-UK Mutation Explorer portal (http://sars2.cvr.gla.ac.uk/cog-uk/, ‘Antigenic Mutations’ tab) and well-ranked journals (such as Cell, Nature, Science, or JAMA). From such publications, we extracted relevant information concerning epidemiological effects (including, e.g., viral transmission, infectivity, virulence); immunological effects (including, e.g., sensitivity to monoclonal antibodies and binding affinity to hosts’ receptors); or protein kinetics effects (such as protein flexibility and stability). The presence of the change may yield an increase or decrease of the impact (indicated with the H=higher or L=lower labels); effects are usually reported as a result of a scientific study employing an epidemiological, experimental, or computational method. A detailed list of the considered effects can be found at https://github.com/DEIB-GECO/cov2k_data_collector/blob/master/CoV2K_Effects_Taxonomy.pdf. In some cases, an effect may be connected to a single amino acid change or to a specific group of changes.
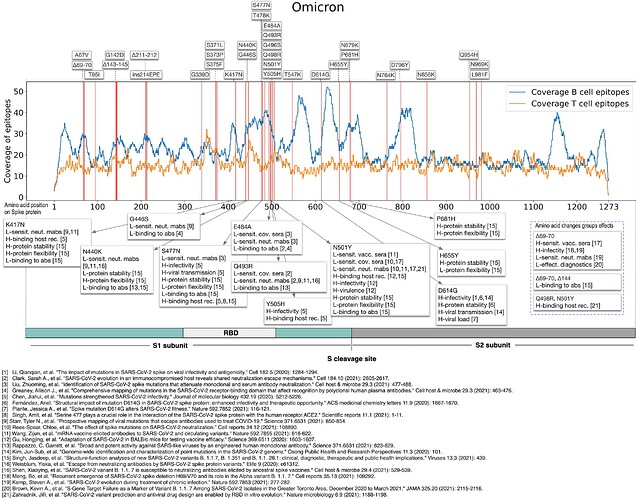
Finally, the above figure summarizes, along the Spike protein, all Omicron mutated positions (according to CoVariants list) and annotates some of the mutations with the effects found as described above. The notation “L-sensit. neut. mabs [9,11]” assigned to K417N means that two literature references (9 and 11) have reported that K417N lowers the sensitivity of the virus with respect to neutralizing monoclonal antibodies. Some effects have been found for the groups of deletions on positions 69 and 70 (possibly with the addition of the deletion on position 144) and for the co-occurring substitutions on positions 498 and 501. A relevant number of papers had previously discussed effects caused by the co-occurring mutations E484K, Q498R, and N501Y [C, D], but this does not apply to Omicron as the 484th residue E (Glutamic acid) is changed to A (Alanine) rather than to K (Lysine).
[A] Vita, Randi, et al. “The immune epitope database (IEDB): 2018 update.” Nucleic acids research 47.D1 (2019): D339-D343.
[B] Bernasconi, Anna, et al. “EpiSurf: metadata-driven search server for analyzing amino acid changes within epitopes of SARS-CoV-2 and other viral species.” Database 2021 (2021).
[C] Zahradník, Jiří, et al. “SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution.” Nature microbiology 6.9 (2021): 1188-1198.
[D] Zhou, Daming, et al. “Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera.” Cell 184.9 (2021): 2348-2361.