Preliminary in silico assessment of the specificity of published molecular assays and design of new assays using the available whole genome sequences of 2019-nCoV
Authors:
Mitchell Holland1, Daniel Negrón1, Shane Mitchell1, Mychal Ivancich1, Katharine W. Jennings1, Bruce Goodwin2, and Shanmuga Sozhamannan2.3
1Noblis, Reston, VA 20191
2Defense Biological Product Assurance Office, Frederick, MD 21702
3Logistics Management Institute, Tysons, VA 22102
Acknowledgement: All the investigators/labs that published the sequence data at GISAID- also see table at the end
Similar analyses/References: Marion Koopmans; Initial assessment of the ability of published coronavirus primers sets to detect the Wuhan coronavirus: http://virological.org/t/initial-assessment-of-the-ability-of-published-coronavirus-primers-sets-to-detect-the-wuhan-coronavirus/321Source
WGS: 25 WGS sequences have been downloaded from GISAID; All other data from NCBI BLAST databases (nt, gss, and env_nt - last updated December 2019)
Attached zip file with data visualizations: nCoV_pset_report.zip (459.5 KB)
Analyses:
BioLaboro is an application for rapidly designing de novo assays and validating existing PCR detection assays. It is a user-friendly new assay discovery pipeline composed of three tools: BioVelocity®, Primer3, and PSET. BioVelocity® uses a rapid, accurate hashing algorithm to align sequencing reads to a large set of references (e.g. Genbank) (Sozhamannan et al., 2015). BioVelocity® creates a k-mer index to determine all possible matches between query sequences and references simultaneously using a large RAM system (i.e. an IBM Power8). This algorithm makes it possible to very quickly identify sequences conserved within or omitted from a set of target references. Primer3 (http://primer3.sourceforge.net/) is a tool for designing primers and probes for real-time PCR reactions. It considers a range of criteria such as oligonucleotide melting temperature, size, GC content, and primer-dimer possibilities. We use Primer3 along with our signature detection process to identify potential new primer sets. PSET (PCR Signature Erosion Tool) tests PCR assays in silico against the latest versions of public sequence repositories, or other reference datasets, to determine if primers and probes match only to their intended targets. Using this information, an assay provider can be better aware of potential false hits and be better prepared to design new primers when false hits become intractable.
Results:
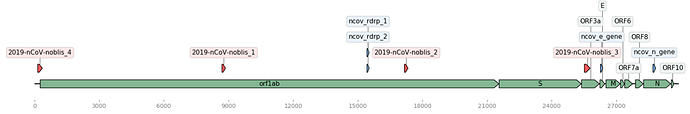
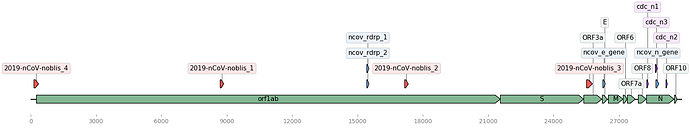
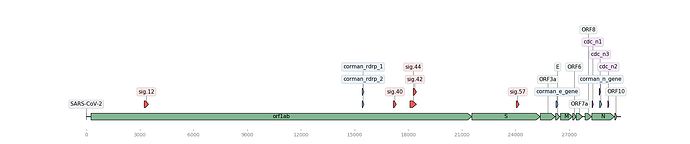
The BioLaboro application detected four highly specific signature sequence regions that hit all 25 (available at the time of analysis) Wuhan genomes (Table 1). The detected signatures were found to occur in disparate locations on the genome (Figure 1). All four signatures were found to target all current Wuhan genomes, and three out of four of these signature regions did not sufficiently align to any known coronavirus or other organism in NCBI BLAST databases (Table 2).
Table 1. List of PCR assays evaluated in this analysis. First four assays newly created using BioLaboro to be specific to Wuhan coronavirus. Last four assays from Diagnostic detection of Wuhan coronavirus 2019 by real-time RTPCR by Corman et al 2020.
| Identifier | length | forward | probe | reverse |
|---|---|---|---|---|
| 2019-nCoV-noblis_1 | 165 | TGATGGTGGTGTCACTCGTG | TGGTTTAGCCAGCGTGGTGGT | GAAGTGGGTTTTGTCGTGCC |
| 2019-nCoV-noblis_2 | 168 | GCCGCTGTTGATGCACTATG | ACGTGCTCGTGTAGAGTGTTTTGAT | ATGCATTGCCTGAGACGACA |
| 2019-nCoV-noblis_3 | 272 | CGGATGGCTTATTGTTGGCG | TGCTCGTTGCTGCTGGCCTT | TTGGCTTTGCTGGAAATGCC |
| 2019-nCoV-noblis_4 | 218 | TGTCGTTGACAGGACACGAG | TTCGTCCGTGTTGCAGCCGA | CGTACGTGGCTTTGGAGACT |
| ncov_e_gene | 113 | ACAGGTACGTTAATAGTTAATAGCGT | ACACTAGCCATCCTTACTGCGCTTCG | TGTGTGCGTACTGCTGCAATAT |
| ncov_n_gene | 128 | CACATTGGCACCCGCAATC | ACTTCCTCAAGGAACAACATTGCCA | CAAGCCTCTTCTCGTTCCTC |
| ncov_rdrp_1 | 100 | GTGARATGGTCATGTGTGGCGG | CCAGGTGGWACRTCATCMGGTGATGC | TATGCTAATAGTGTSTTTAACATYTG |
| ncov_rdrp_2 | 100 | GTGARATGGTCATGTGTGGCGG | CAGGTGGAACCTCATCAGGAGATGC | TATGCTAATAGTGTSTTTAACATYTG |
Figure 1. Map of the Wuhan genome (NCBI Accession: MN908947.3) with assay signature locations (created using DNA Features Viewer Python library). Corman assays in blue, Noblis assays in red, and gene regions in green.
Table 2. Results from PSET analysis. The four new Noblis assays were compared alongside the four assays from Corman. Each assay was tested using Wuhan coronavirus (25 genomes) as the intended target. All off-target hits (PF, FP) are to entries in NCBI BLAST databases (nt, gss, and env_nt).
| Assay | Confusion | |||||||
|---|---|---|---|---|---|---|---|---|
| identifier | type | target | PT | TP | TN | PF | FP | FN |
| 2019-nCoV-noblis_1 | Probe | Wuhan | 22 | 3 | 415 | NA | NA | NA |
| 2019-nCoV-noblis_2 | Probe | Wuhan | 25 | NA | 691 | NA | NA | NA |
| 2019-nCoV-noblis_3 | Probe | Wuhan | 25 | NA | 656 | NA | NA | NA |
| 2019-nCoV-noblis_4 | Probe | Wuhan | 25 | NA | 356 | NA | 3 | NA |
| ncov_e_gene | Probe | Wuhan | 25 | NA | 75 | 353 | 15 | NA |
| ncov_n_gene | Probe | Wuhan | 25 | NA | 73 | NA | 339 | NA |
| ncov_rdrp_1 | Probe | Wuhan | NA* | 25 | 169 | 433 | 85 | NA |
| ncov_rdrp_2 | Probe | Wuhan | NA* | 25 | 586 | 1 | 61 | NA |
| PT = Perfect True Positive = All assay components hit with 100% identity to the correct target | ||||||||
| TP = True Positive = All assay components hit with >=90% identity over >=90% of the component length to the correct target | ||||||||
| TN = True Negative = Partial hit to assay amplicon but insufficient assay component alignments to an incorrect target | ||||||||
| PF = Perfect False Positive = All assay components hit with 100% identity to an incorrect target | ||||||||
| FP = False Positive = All assay components hit with >=90% identity over >=90% of the component length to an incorrect target | ||||||||
| FN = False Negative = Partial hit to assay amplicon but insufficient assay component alignments to the correct target | ||||||||
| * The ncov_rdrp_1 and ncov_rdrp_2 assays have one mismatch in the reverse primer to all current Wuhan coronavirus genomes which prevents it from being called a perfect hit. This is an incorrect match between an ambiguous base code, “S”, and the reference sequence “T”, in the middle of the primer which will likely not affect binding. NA == 0. |
Conclusions and Caveats:
This is preliminary analyses based on 25 sequences available at the time.
New sequences are added on an hourly basis and these Noblis signatures need to be tested against the new sequences to verify that no signature erosion is occurring, as described in Sozhamannan et al 2015 for Ebola sequences. These designs were generated entirely in silico , and have yet to be tested in the lab. Although the BioLaboro pipeline is designed on sound scientific principles and the results from analyses of Ebola and Lassa viruses using the in silico components have been demonstrated (Sozhamannan et al 2015 and Wiley et al 2019) these assays await validation before conclusions regarding their use for clinical testing can be made.
Our intent in publishing these nCoV real-time PCR assays is to make the community aware of the existence of these potential unique signature regions as well the availability of BioLaboro for rapid evaluation of existing assays and design of new assays.
References:
-
Diagnostic detection of Wuhan coronavirus 2019 by real-time RTPCR -Protocol and preliminary evaluation as of Jan 13, 2020- Victor Corman, Tobias Bleicker, Sebastian Brünink, Christian Drosten, Charité Virology, Berlin, Germany; Olfert Landt, Tib-Molbiol, Berlin, Germany; Marion Koopmans, Erasmus MC, Rotterdam, The Netherlands; Maria Zambon, Public Health England, London, Additional advice by Malik Peiris, University of Hong Kong; contact: [email protected] Institute of Virology - Charité – Universitätsmedizin Berlin
-
Sozhamannan, Shanmuga, et al. “Evaluation of signature erosion in Ebola virus due to genomic drift and its impact on the performance of diagnostic assays.” Viruses 7.6 (2015): 3130-3154.
-
Wiley, Michael R., et al. “Lassa virus circulating in Liberia: a retrospective genomic characterisation.” The Lancet Infectious Diseases 19.12 (2019): 1371-1378.
Appendix 1: Acknowledgement for 2019-nCoV genome sequences
The following table is from this blog at Virological.org: Phylogenetic analysis of 23 nCoV-2019 genomes, 2020-01-23; Phylogenetic analysis of 23 nCoV-2019 genomes, 2020-01-23.
Table 3 . nCoV2019 genome sequences used in this analysis, the GISAID 6 accession numbers and submitting labs.
| GISAID Accession | Strain | Location | Collection date | Lab |
|---|---|---|---|---|
| EPI_ISL_404227 | BetaCoV/Zhejiang/WZ-01/2020 | Zhejiang, China | 2020-01-16 | 1 |
| EPI_ISL_404228 | BetaCoV/Zhejiang/WZ-02/2020 | Zhejiang, China | 2020-01-17 | 1 |
| EPI_ISL_402132 | BetaCoV/Wuhan/HBCDC-HB-01/2019 | China/Hubei Province | 2019-12-30 | 2 |
| EPI_ISL_402127 | BetaCoV/Wuhan/WIV02/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 3 |
| EPI_ISL_402128 | BetaCoV/Wuhan/WIV05/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 3 |
| EPI_ISL_402129 | BetaCoV/Wuhan/WIV06/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 3 |
| EPI_ISL_402130 | BetaCoV/Wuhan/WIV07/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 3 |
| EPI_ISL_403963 | BetaCoV/Nonthaburi/74/2020 | Thailand/ Nonthaburi Province | 2020-01-13 | 4 |
| EPI_ISL_403962 | BetaCoV/Nonthaburi/61/2020 | Thailand/ Nonthaburi Province | 2020-01-08 | 4 |
| EPI_ISL_402120 | BetaCoV/Wuhan/IVDC-HB-04/2020 | China / Hubei Province / Wuhan City | 2020-01-01 | 5 |
| EPI_ISL_402119 | BetaCoV/Wuhan/IVDC-HB-01/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 5 |
| EPI_ISL_402121 | BetaCoV/Wuhan/IVDC-HB-05/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 5 |
| EPI_ISL_402124 | BetaCoV/Wuhan/WIV04/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 6 |
| EPI_ISL_402123 | BetaCoV/Wuhan/IPBCAMS-WH-01/2019 | China / Hubei Province / Wuhan City | 2019-12-24 | 7 |
| EPI_ISL_402125 | BetaCoV/Wuhan-Hu-1/2019 | China | 2019-12 | 8 |
| EPI_ISL_403931 | BetaCoV/Wuhan/IPBCAMS-WH-02/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 9 |
| EPI_ISL_403928 | BetaCoV/Wuhan/IPBCAMS-WH-05/2020 | China / Hubei Province / Wuhan City | 2020-01-01 | 9 |
| EPI_ISL_403930 | BetaCoV/Wuhan/IPBCAMS-WH-03/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 9 |
| EPI_ISL_403929 | BetaCoV/Wuhan/IPBCAMS-WH-04/2019 | China / Hubei Province / Wuhan City | 2019-12-30 | 9 |
| EPI_ISL_403937 | BetaCoV/Guangdong/20SF040/2020 | Guangdong, China | 2020-01-18 | 10 |
| EPI_ISL_403936 | BetaCoV/Guangdong/20SF028/2020 | Guangdong, China | 2020-01-17 | 10 |
| EPI_ISL_403935 | BetaCoV/Guangdong/20SF025/2020 | Guangdong, China | 2020-01-15 | 10 |
| EPI_ISL_403934 | BetaCoV/Guangdong/20SF014/2020 | Guangdong, China | 2020-01-15 | 10 |
| EPI_ISL_403933 | BetaCoV/Guangdong/20SF013/2020 | Guangdong, China | 2020-01-15 | 10 |
| EPI_ISL_403932 | BetaCoV/Guangdong/20SF012/2020 | Guangdong, China | 2020-01-14 | 10 |
[1] Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention
[2] Hubei Provincial Center for Disease Control and Prevention
[3] Wuhan Institute of Virology, Chinese Academy of Sciences
[4] Department of Medical Sciences, Ministry of Public Health, Thailand & Thai Red Cross Emerging Infectious Diseases - Health Science Centre & Department of Disease Control, Ministry of Public Health, Thailand
[5] National Institute for Viral Disease Control and Prevention, China CDC
[6] Wuhan Institute of Virology, Chinese Academy of Sciences
[7] Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College
[8] National Institute for Communicable Disease Control and Prevention (ICDC) Chinese Center for Disease Control and Prevention (China CDC)
[9] Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College
[10] Department of Microbiology, Guangdong Provincial Center for Diseases Control and Prevention