First report of COVID-19 in Scotland
We report the genomic analysis of the first positive SARS-CoV-2 sample retrieved from a patient in Scotland on 2nd March 2020. Following detection, a sample was processed by the West of Scotland Specialist Virology Centre, NHSGGC and sent to the MRC-University of Glasgow Centre for Virus Research (CVR) for sequencing.
A confirmation diagnostic was conducted using real-time RT-PCR protocol to detect SARS-CoV-2. RT-PCR results had a cycle threshold value of 32 and 36, depending on the protocol used. Nucleic acid was extracted utilising a NucliSENS® easyMag® platform followed by mild DNase treatment. Sequencing was performed on Oxford Nanopore Technologies MinION sequencer utilising protocols developed by the ARTIC network where the sequencing protocol and multiplex primers are described in detail. Library preparation was conducted without a barcoding step and 50 fmol of the sequencing library was loaded onto the flow cell (R9.4.1), according to standard protocol. Sequencing was conducted in MinKNOW version 19.12.5 for 4 hours. A total of 1,501,467 reads were generated. Raw FAST5 files were basecalled with Guppy (version 3.4.5) in high accuracy mode and using a minimum quality score of 7. The ARTIC nCoV-2019 environment utilising RAMPART (v1.0.5) was used to assign and map reads in real-time. Following the ARTIC-nCov-2019 bioinformatics protocol, reads were initially size filtered, then demultiplexed and trimmed with Porechop, and mapped against reference strain Wuhan-Hu-1 (GenBank accession number MN908947), followed by clipping of primer regions. 98% of the genome was covered with a depth of >20 reads. Variants were called using nanopolish 0.11.3 and accepted if they had a log-likelihood score of greater than 200. In the resulting consensus sequence, low coverage regions are masked with N characters.
The genome was named Scotland/CVR01/2020 and has been deposited in GISAID (EPI_ISL_413221).
To determine the epidemiological linkage to circulating SARS-CoV-2 variants, the genome was uploaded to our CoV-GLUE resource. This analysis identified nine nucleotide and five amino acid changes compared to the Wuhan-Hu-1 genome (nsp12:P323L, S:D614G, N:R203K and N:G204R, ORF7a:T120I), only one of which is novel to this virus (ORF7a:T120I). To infer an evolutionary tree CVR01 was included in an alignment of global SARS-CoV-2 sequences using MAFFT, checked with Geneious and a maximum likelihood phylogenetic tree constructed using PhyML with a GTR nucleotide substitution model and gamma rate heterogeneity model.
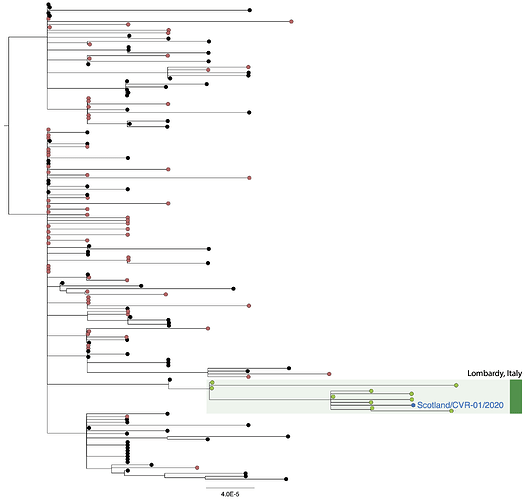
Figure 1 | Phylogenetic tree showing relationship of CVR01 to other SARS-CoV-2 genomes. Sequences associated with travel to northern Italy are indicated in green. The scale bar indicates substitutions per nucleotide site.
In phylogenetic analysis CVR01 clusters with viruses from individuals from Mexico, Germany and Switzerland who had recently travelled to Italy, indicating no evidence of undetected SARS-CoV-2 circulating in Scotland. Surveillance is ongoing in Scotland and the UK to monitor for SARS-CoV-2 spread.
This is a collaboration between the MRC-University of Glasgow Centre for Virus Research (CVR), West of Scotland Specialist Virology Centre, NHSGGC and University of Edinburgh, and is part of the ISARIC-Coronavirus Clinical Characterisation Consortium (ISARIC-4C) consortium headed by Kenneth Baillie, Roslin Institute, University of Edinburgh; Calum Semple, University of Liverpool; and Peter Openshaw, Imperial College London.
Contributors: CVR clinical researchers: Emma Thomson & Antonia Ho; CVR Thomson lab: James Shephard & Shirin Ashraf; CVR Genomics: Kathy Smollett, Daniel Mair, Stephen Carmichael & Ana da Silva Filipe; CVR Bioinformatics: Richard Orton, Josh Singer, Scott Arkison & David L Robertson; University of Edinburgh and Artic Network: Andrew Rambaut; WoSSVC, NHSGGC: Alasdair MacLean and Rory Gunson.
Acknowledgments
We would like to thank all the authors who have kindly deposited and shared genome data on GISAID. A table with genome sequence acknowledgments can be found on the CoV-GLUE website. Thanks to Josh Quick and Nick Loman, University of Birmingham, for providing ARTIC primers.