We report the genomic analysis of four SARS-CoV-2/hCOV-19 samples retrieved from patients in Scotland, in early March 2020. Following detection, samples were processed by the West of Scotland Specialist Virology Centre, NHSGGC and sent to the MRC-University of Glasgow Centre for Virus Research (CVR) for library preparation and sequencing. See our First report of COVID-19 in Scotland for details on the Oxford Nanopore Technologies MinION sequencing protocol utilised.
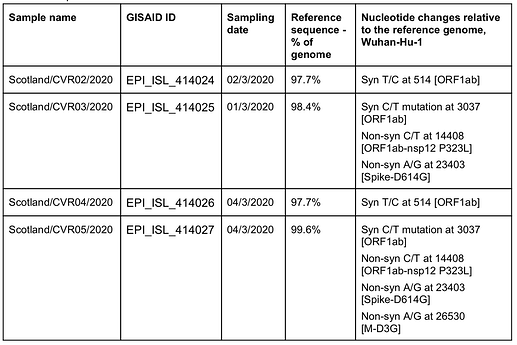
The four samples were named CVR02 to CVR05 and have been deposited in GISAID (see table 1 for IDs, date of sampling, coverage and nucleotide changes).
Table 1. Sample data.
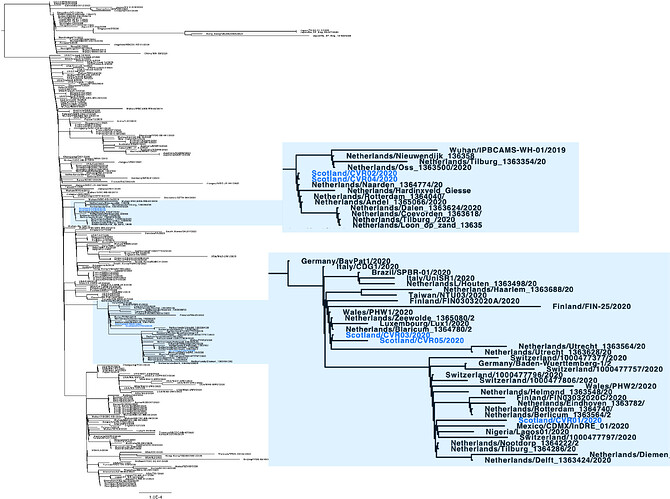
To determine their relationship to circulating SARS-CoV-2/hCOV-19 variants, an evolutionary tree was inferred using these genomes and all complete human genomes available from GISAID; a small number of GISAID sequences were discounted due to large number of mutations, presumably from sequence errors. Briefly, an alignment was generated using MAFFT and a maximum likelihood phylogenetic tree constructed using RAxML with a GTR nucleotide substitution model and gamma rate heterogeneity model. The tree was visualised with FigTree and mid-point rooted.
Figure 1 | Phylogenetic tree showing the relationship of CVR01-05 to other SARS-CoV-2/hCoV-19 genomes. The Scottish sequences are highlighted in blue and in zoomed views on the right. The scale bar indicates substitutions per nucleotide site.
Including CVR01, our analysis indicates three introductions of COVID-19 infections into Scotland for which genome sequence data is available. For two of these there is evidence of linked transmissions either occurring in or out of Scotland: (1) CVR02 and CVR04 are identical so cluster together and (2) CVR03 and CVR05 also cluster together, CVR05 has one additional mutation. Clustering is consistent with the known travel histories.
It is important to note the countries indicated in the sequence names are where the individuals reside and not necessarily the origin of their infection, as most are recent travellers. For example, CVR02 and CVR04 are closest to samples from individuals testing positive in the Netherlands. However, it cannot be concluded from this analysis that the Scottish individuals were infected in the Netherlands as we do not have complete travel histories for these cases.
Importantly, there are now multiple introductions of SARS-CoV-2/hCoV-19 into Europe and several introductions to Scotland. A tracker of UK case counts has been setup by PHE.
Contributors: CVR clinical researchers1: Emma Thomson & Antonia Ho; CVR Genomics1: Kathy Smollett, Daniel Mair, Stephen Carmichael & Ana da Silva Filipe; CVR Bioinformatics1: Richard Orton & David L Robertson; WoSSVC, NHSGGC2: Alasdair MacLean and Rory Gunson.
1MRC-University of Glasgow Centre for Virus Research (CVR); 2West of Scotland Specialist Virology Centre, NHSGGC and University of Edinburgh.
Acknowledgments
We would like to thank all the authors who have kindly deposited and shared genome data on GISAID. A table with genome sequence acknowledgments can be found on the CoV-GLUE.