Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2
Áine O’Toole1,$,*, Verity Hill1*, Oliver G. Pybus2, Alexander Watts3,4, Isaac I. Bogoch5,6, Kamran Khan3,4,5, Jane P. Messina7, The COVID-19 Genomics UK (COG-UK) consortium8, Network for Genomic Surveillance in South Africa (NGS-SA)9, Brazil-UK CADDE Genomic Network10, Houriiyah Tegally11, Richard R Lessells11, Jennifer Giandhari11, Sureshnee Pillay11, Kefentse Arnold Tumedi13, Gape Nyepetsi14, Malebogo Kebabonye15, Maitshwarelo Matsheka13, Madisa Mine14, Sima Tokajian16, Hamad Hassan17, Tamara Salloum18, Georgi Merhi18, Jad Koweyes18, Jemma L Geoghegan19,20, Joep de Ligt20, Xiaoyun Ren20, Matthew Storey20, Nikki E Freed21,Chitra Pattabiraman22, Pramada Prasad22, Anita S Desai22, Ravi Vasanthapuram22,Thomas F. Schulz23, Lars Steinbrück23, Tanja Stadler24, Swiss Viollier Sequencing Consortium25, Antonio Parisi26, Angelica Bianco26, Darío García de Viedma27,28,29, Sergio Buenestado-Serrano27,28, Vítor Borges30, Joana Isidro30, Sílvia Duarte31, João Paulo Gomes30, Neta S. Zuckerman32, Michal Mandelboim32, Orna Mor32, Torsten Seemann33, Alicia Arnott34, Jenny Draper34, Mailie Gall34, William Rawlinson35, Ira Deveson36, Sanmarié Schlebusch37, Jamie McMahon37, Lex Leong38, Chuan Kok Lim38,Maria Chironna39, Daniela Laconsole39, Antonin Bal40, Laurence Josset40, Edward Holmes41, Kirsten St George42, Erica Lasek-Nesselquist42, Reina S. Sikkema43, Bas B. Oude Munnink43, Marion Koopmans43, Mia Brytting44, V. Sudha Rani45, S. Pavani45, Teemu Smura46, Albert Heim47, Satu Kurkela48, Massab Umair52, Muhammad Salman52, Barbara Bartolini53, Martina Rueca53, Christian Drosten54, Thorsten Wolff56, Olin Silander21, Dirk Eggink59, Chantal Reusken59, Harry Vennema59, Aekyung Park60, SEARCH Alliance San Diego48, National Virus Reference Laboratory49, SeqCOVID-Spain50, Danish Covid-19 Genome Consortium (DCGC)55, Communicable Diseases Genomic Network (CDGN)57, Dutch National SARS-CoV-2 surveillance program58,#, Division of Emerging Infectious Diseases KDCA60, Tulio de Oliveira11, Nuno R. Faria2,12, Andrew Rambaut1, Moritz U. G. Kraemer2,$
Affiliations:
- Institute of Evolutionary Biology, University of Edinburgh, United Kingdom
- Department of Zoology, University of Oxford, United Kingdom
- Li Ka Shing Knowledge Institute, St. Michael’s Hospital, Toronto, Canada
- BlueDot, Toronto, Canada
- Department of Medicine, University of Toronto, Toronto, Canada
- Divisions of General Internal Medicine and Infectious Diseases, University Health Network, Toronto, Canada
- Department of Geography, University of Oxford, United Kingdom
- https://www.cogconsortium.uk/
- Kwazulu-Natal Research Innovation and Sequencing Platform
- Brazil-UK CADDE Genomic Network
- KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP), Nelson R Mandela School of Medicine, University of KwaZulu-Natal, Durban, South Africa
- Imperial College London, United Kingdom
- Botswana Institute for Technology Research and Innovation, Gaborone, Botswana
- National Health Laboratory, Gaborone, Botswana
- Ministry of Health and Wellness, Botswana
- Department of Natural Sciences, Lebanese American University
- Faculty of Public Health, Lebanese University
- Department of Natural Sciences, Lebanese American University
- Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
- Institute of Environmental Science and Research, Wellington, New Zealand
- School of Natural and Computational Sciences, Massey University, Auckland, New Zealand.
- Department of Neurovirology, National Institute of Mental Health and Neurosciences, Bengaluru, India
- Institute of Virology, Hannover Medical School, Hannover, Germany
- Department of Biosystems Science and Engineering, ETH Zürich, Switzerland
- Swiss SARS-CoV-2 Sequencing Consortium (S3C) – Computational Evolution | ETH Zurich
- Istituto Zooprofilattico sperimentale della Puglia e della Basilicata
- Hospital General Universitario Gregorio Marañón;
- Instituto de Investigación Sanitaria Gregorio Marañón, Madrid. Spain.
- CIBER Enfermedades Respiratorias CIBERES, Spain.
- Bioinformatics Unit, Department of Infectious Diseases, National Institute of Health Doutor Ricardo Jorge (INSA), Lisbon, Portugal
- Innovation and Technology Unit, Department of Human Genetics, National Institute of Health Doutor Ricardo Jorge (INSA), Lisbon, Portugal
- Central Virology Laboratory, Israel Ministry of Health, Sheba Medical Center, Israel
- Microbiological Diagnostic Unit Public Health Laboratory, Department of Microbiology & Immunology, University of Melbourne at the Peter Doherty Institute for Infection & Immunity, Melbourne, Victoria, Australia
- New South Wales Health Pathology - Institute of Clinical Pathology and Medical Research, Sydney, New South Wales, Australia
- New South Wales Health Pathology Randwick, Prince of Wales Hospital, Sydney, New South Wales, Australia
- Kinghorn Centre for Clinical Genomics, Sydney, New South Wales, Australia
- Queensland Reference Centre for Microbial and Public Health Genomics, Forensic and Scientific Services, Health Support Queensland, Queensland Health
- South Australia Pathology, Adelaide, South Australia, Australia
- Department of Biomedical Sciences and Human Oncology, University of Bari
- Centre National de Référence des virus des infections respiratoires, Hospices Civils de Lyon, Lyon, France
- University of Sydney, Sydney, New South Wales, Australia
- Wadsworth Center, New York State Department of Health, Albany, New York
- ErasmusMC, Department of Viroscience, WHO collaborating centre for arbovirus and viral hemorrhagic fever Reference and Research, Rotterdam, the Netherlands
- The Public Health Agency of Sweden, Department of Microbiology, Solna, Sweden
- Upgraded Department of Microbiology, Osmania Medical College, Hyderabad, Telangana, India
- Department of Virology, University of Helsinki, Helsinki, Finland
- Institute of Virology, Hannover Medical School, Hannover, Germany
- The Scripps Research Institute, La Jolla, California
- National Virus Reference Laboratory, University College Dublin, Belfield, Dublin, Republic of Ireland
- http://seqcovid.csic.es/
- HUS Diagnostic Center, HUSLAB, Clinical Microbiology, University of Helsinki and Helsinki University Hospital, Finland
- Department of Virology, National Institute of Health, Pakistan
- National Institute for Infectious Diseases “L. Spallanzani”, Via Portuense, 292, 00149 Rome, Italy
- Institute for Virology, Charité Universitätsmedizin, Berlin, Germany
- https://www.covid19genomics.dk/
- Robert Koch-Institut, Head, Unit 17, Influenza and other Respiratory Viruses, Seestr. 10, Berlin, Germany
- https://www.cdgn.org.au/
- National Coordination Centre for Communicable Disease Control | RIVM
- WHO COVID-19 reference laboratory, Centre for Infectious Disease Control-National Institute for Public Health and the Environment, Bilthoven the Netherlands
- Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency
*contributed equally
$correspondence should be addressed to [email protected] or [email protected]
# Full list of consortium names and affiliations are in the Supplementary Materials
Latest data January 7, 2020

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
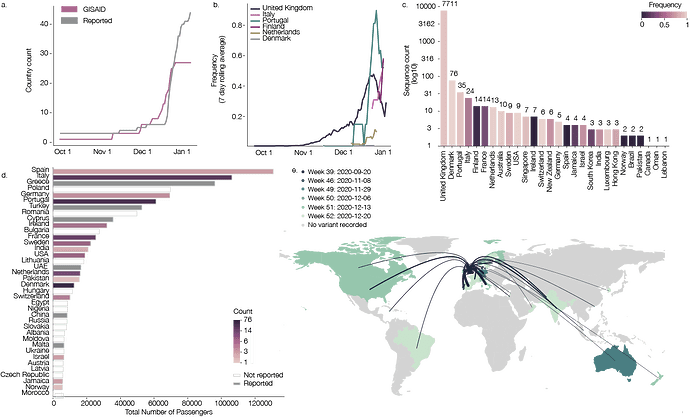
In December 2020, routine genomic surveillance in the United Kingdom (UK)11 reported a new and genetically-distinct phylogenetic cluster of SARS-CoV-2 (variant VOC202012/01, lineage B.1.1.7). Preliminary analysis suggests that this lineage carries an unusually large number of genetic changes10. The earliest known cases of B.1.1.7 were sampled in southern England in late September 2020; by December the lineage had spread to most UK regions and was growing rapidly9. In October 2020, a separate SARS-CoV-2 cluster (variant 501Y.V2, lineage B.1.351), which carried a different constellation of genetic changes, was detected by the Network for Genomic Surveillance in South Africa7,8. Both lineages carry mutations, especially in the virus spike protein, that may affect virus function, and both appear to have grown rapidly in relative frequency since their discovery. Early analyses of the spatial spread of SARS-CoV-2 highlights the potential for rapid virus dissemination through national and international travel1,6. Therefore continued genomic monitoring of lineages of concern is required.
To better characterise the international distribution of lineages B.1.1.7 and B.1.351 we collated SARS-CoV-2 sequences from GISAID (gisaid.org) and assigned lineages using pangolin (v2.1.6, GitHub - cov-lineages/pangolin: Software package for assigning SARS-CoV-2 genome sequences to global lineages.), which implements the nomenclature scheme described in Rambaut et al.,5. Genomes are assigned lineage to B.1.1.7 if they exhibit at least 5 of the 17 mutations inferred to have arisen on the phylogenetic branch immediately ancestral to the cluster (Supplementary table 1)10; or to B.1.351 if they exhibit at least 5 of 9 lineage-associated mutations (Supplementary table 1)8. Lineage count and frequency data have been calculated daily and are presented online at Redirecting….
As of 7th Jan 2021, 45 countries had reported the presence of B.1.1.7 and 13 countries had reported B.1.351/501Y.V2. B.1.1.7 and B.1.351 genome sequences were available for 28 and 8 countries, respectively (Figure 1a, b, c). Although some countries report increases in the relative frequency of B.1.1.7, genome sequencing efforts vary considerably. Potential targeting of sequencing towards travellers from the UK could bias frequency estimates upwards (Figure 1b, c) and differing genome deposition policies and delays may also skew reporting estimates. The time between the initial collection date of a new variant sample in a country and the first availability of a corresponding virus genome on GISAID was, on average, 12 days (range 1-71).
The number of B.1.1.7 and B.1.351/501Y.V2 genome sequences reported in each country is a consequence of (i) the intensity of local genomic surveillance; (ii) the level of concern about new variant introductions; (iii) the volume of international travel among affected countries, and (iv) the amount of local transmission following the introduction of lineage from elsewhere. To explore these factors, we analysed the most recent available International Air Transport Association (IATA) travel data (October 2020). We collated the total number of origin-to-destination air journeys between major London international airports and each country. The calculation was repeated for journeys originating in all international South African airports. We focussed on London and South Africa as they are the locations with the first reports and highest reported prevalences of lineages B.1.1.7 and B.1.351 respectively 8,10. However, due to low SARS-CoV-2 genomic surveillance in many locations, we cannot reject the hypotheses that these lineages initially originated elsewhere. Figure 1d shows destinations receiving >5,000 travellers in October 2020 from the UK (Supplementary Figure 1 shows destinations receiving >300 travellers from South Africa).
Of the countries that receive >5,000 travellers from London, 16 have sequenced B.1.1.7. Of the 45 countries that have identified B.1.1.7 (32 in travellers and 13 with local onward transmission), only 6 perform real-time routine genomic surveillance (Denmark, UK, Iceland, The Netherlands, Australia, Sweden), 3 have prioritised sequencing based on S-gene target failure tests 4, 30 primarily targeted sequencing towards arriving travellers from the UK, and no information was available for 10 countries (details at lineages-website/_data at master · cov-lineages/lineages-website · GitHub). Of the 13 countries that have identified B.1.351 (four with local onward transmission including South Africa), 4 perform routine sequencing (South Africa, UK, Botswana, Australia), 6 target sequencing of travellers, and no information was available for 3 countries. Consequently, the number of sequences reported does not correlate with flight numbers, but rather reflects current genomic surveillance effort. For example, in September, the UK sequenced ~13% of its reported cases and Denmark sequenced ~21%. In comparison, Israel sequenced ~0.002% of its cases during the same period 2,3.
Our study has several limitations. The passenger flight data do not include recent changes to holiday travel, and recent restrictions on travel from the UK and South Africa is not reflected in the mobility data. Further, flight data may not accurately reflect the final destination if multiple tickets are purchased.
The discovery and rapid spread of B.1.1.7 and B.1.351/501Y.V2 highlights the importance of real-time and open data for tracking the spread of SARS-CoV-2 and for informing future public health interventions and travel advice.
Figure 1: a) The cumulative number of countries with reports of lineage B.1.1.7 (grey line) and cumulative number of genomes of B.1.1.7 deposited in GISAID. b) Rolling seven day average of the proportion of B.1.1.7 genomes in countries with more than ten sequences of the variant, and with more than ten days between the first B.1.1.7 sequence and the most recent one compared to all sampled genomes in that country. c) Number of sequences (log10) per country. Colour indicates the proportion of sequences that are classified as lineage B.1.1.7. d) Number of air travellers from major international London airports (Heathrow, Gatwick, Luton, City, Stansted, Southend) during October 2020. Colour indicates the number of sampled genomes of lineage B.1.1.7. e) Map of international flights from major international London Airports to countries with B.1.1.7 sequences. Colours indicate the date of earliest detection of B.1.1.7. in each country. The width of the lines indicates the number of flights. International Air Transport Association data used here account for ~90% of passenger travel itineraries on commercial flights, excluding transportation via unscheduled charter flights (the remainder is modelled using market intelligence). Data shown represents origin-destination journeys during October 2020. Routes to countries that have not yet detected B.1.1.7 and deposited data on GISAID are not included.
References
1 du Plessis L, McCrone JT, Zarebski AE, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021; published online Jan 8. DOI:10.1126/science.abf2946.
2 Hasell J, Mathieu E, Beltekian D, et al. A cross-country database of COVID-19 testing. Sci Data 2020; 7: 345.
3 Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20: 533–4.
4 Bal A, Destras G, Gaymard A, et al. Two-step strategy for the identification of SARS-CoV-2 variants co-occurring with spike deletion H69-V70, Lyon, France, August to December 2020. bioRxiv. 2020; published online Nov 13. DOI:10.1101/2020.11.10.20228528.
5 Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5: 1403–7.
6 Lu J, du Plessis L, Liu Z, et al. Genomic Epidemiology of SARS-CoV-2 in Guangdong Province, China. Cell 2020; 181: 997–1003.e9.
7 Msomi N, Mlisana K, de Oliveira T, Network for Genomic Surveillance in South Africa writing group. A genomics network established to respond rapidly to public health threats in South Africa. Lancet Microbe 2020; 1: e229–30.
8 Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv. 2020; published online Dec 22. DOI:10.1101/2020.12.21.20248640.
9 Volz E, Mishra S, Chand M, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. bioRxiv. 2021; published online Jan 4. DOI:10.1101/2020.12.30.20249034.
10 Rambaut et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020; published online Dec 18. https://pando.tools/t/563 (accessed Jan 8, 2021).
11 COVID-19 Genomics UK (COG-UK) [email protected]. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe 2020; 1: e99–100.
Funding
I.I.B. is supported by the Canadian Institutes of Health Research, COVID-19 Rapid Research Funding Opportunity (02179-000). K.K. is the founder of BlueDot, a social enterprise that develops digital technologies for public health. K.K., A.W., A.T.B. and C.H. are employed at BlueDot. I.I.B. has consulted for BlueDot. T.d.O. and the NGS-SA is funded by the South African Medical Research Council (SAMRC), MRC SHIP and the Department of Science and Innovation (DSI) of South Africa. N.R.F. acknowledges support from a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (204311/Z/16/Z) and a Medical Research Council-São Paulo Research Foundation CADDE partnership award (MR/S0195/1 and FAPESP 18/14389-0). VH was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [grant number BB/M010996/1]. M.U.G.K. acknowledges support from the Branco Weiss Fellowship and EU grant 874850 MOOD. The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. O.G.P. , J.P.M. and M.U.G.K. acknowledge support from the Oxford Martin School. AR acknowledges the support of the Wellcome Trust (Collaborators Award 206298/Z/17/Z – ARTIC network) and the European Research Council (grant agreement no. 725422 – ReservoirDOCS). A.OT is supported by the Wellcome Trust Hosts, Pathogens & Global Health Programme [grant number: grant.203783/Z/16/Z] and Fast Grants [award number: 2236]. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute.TFS acknowledges support from the Deutsche Forschungsgemeinschaft (SFB900, EXC2155 RESIST). SeqCOVID-SPAIN is supported by a grant from the Instituto de Salud Carlos III COV0020/00140.
Authors contributions
A.OT., O.G.P., J.P.M., N.R.F., A.R., M.U.G.K. conceived the study. A.OT, V.H., M.U.G.K. O.G.P., A.R. wrote the first draft. A.OT, V.H., M.U.G.K., A.R., AW, conducted data analysis. S.T., T.Salloum, G.M., J.K., J.G., J.d.L., X.R., M.S, N.Freed, C.P., P.P., A.D., R.V., T.F.S., L.S., T.Stadler, A.P., A.B., D.G.d.V., S.B-S., V.B., J.I., S.D., J.P.G., N.Z., M.M., O.M., T.Seemann, N.S., B.H., M.Sait, A.A., J.D., M.G., W.R., I.D., S.S., J.M., L.L., C.K.L., M.C., D.L., A.B., L.J., K.S.G., E.L-N., R.S., B.M., M.Koopmans, M.B., V.S.R., S.P. , T.Smura, A.H., S.K., M.U., M.Salman, B.B., M.R., C.D., T.W., O.S., D.E., C.R., H.V., A.P. contributed to the genomic dataset and facilitated data and sample availability. All authors contributed data and contributed to writing.
Acknowledgements
We thank Norelle Sherry, Benjamin Howden and Michelle Sait for their contribution to sequencing in Australia. We acknowledge the work in surveillance and in generating SARS-CoV-2 sequence data by the Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency. We would also like to extend our gratitude to everyone involved in the global sequencing effort.
We also include full acknowledgements in the supplementary materials attached.
Full acknowledgements, consortium author lists and supplemental materials.pdf (373.1 KB)