SARS-CoV-2: don’t ignore non-canonical genes
Zachary Ardern1*, Xinzhu Wei2, Chase W Nelson3*
- Institute for Biological Interfaces 5, Karlsruhe Institute of Technology, Karlsruhe, Germany
- Departments of Computer Science, Human Genetics, and Computational Medicine, University of California, Los Angeles, Los Angeles, CA, USA
- Institute for Comparative Genomics, American Museum of Natural History, New York City, NY, USA
*Correspondence: [email protected]; [email protected]
Non-canonical genes have been largely ignored in emerging viruses. The genomes of viruses closely related to SARS-CoV-2 vary in both accessory and out-of-frame (i.e., overlapping) genes. However, unwarranted methodological assumptions often exclude these genes from consideration, despite their importance for virology, evolution, zoonosis, antigenic potential, vaccines, and therapeutics (Firth and Brierley 2012; Ho et al. 2021; Pavesi 2021).
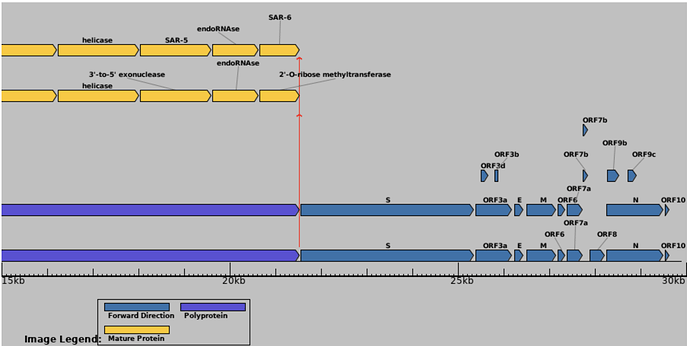
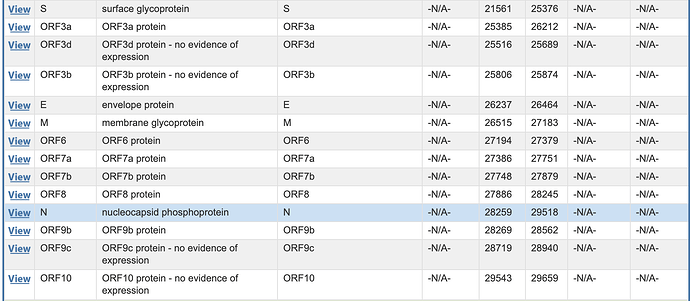
Known or putative non-canonical genes in SARS-CoV-2 include out-of-frame genes ORF2b overlapping Spike; ORF3c, ORF3d, and ORF3b overlapping ORF3a; and ORF9b and ORF9c overlapping N (Firth 2020; Jungreis, Nelson, et al. 2021). Each of these genes shows evidence of translation for one or more isoforms from ribosome profiling (Finkel et al. 2021), mass spectrometry (Zecha et al. 2020), or HLA-I presentation (Nagler et al. 2021; Weingarten-Gabbay et al. 2021).
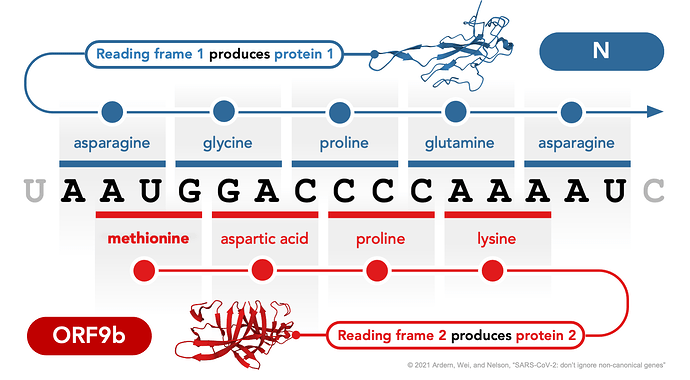
Among out-of-frame genes in SARS-CoV-2, ORF3b is an interferon antagonist (Konno et al. 2020) that elicits a substantial immunoglobulin G (IgG) response (Li et al. 2021) and is dramatically truncated in SARS-CoV-2 compared to SARS-CoV. Another, ORF3d (Nelson, Ardern, Goldberg, et al. 2020), elicits one of the strongest antibody responses observed in patient sera (Hachim et al. 2020), although it has been truncated in some lineages (Jungreis, Sealfon, and Kellis 2021). Confusingly, ORF3d has erroneously been referred to as ORF3b in many studies (documented in Jungreis, Nelson, et al. 2021). A third example, ORF9b (Figure 1), also elicits a strong IgG response (Li et al. 2021) and may contribute to the increased transmissibility of the SARS-CoV-2 Alpha (B.1.1.7) variant (Thorne et al. 2021).
Figure 1 | Translation of proteins N and ORF9b from alternative reading frames of the same locus in the SARS-CoV-2 genome. Non-canonical out-of-frame (i.e., overlapping) genes occur when one nucleotide sequence is translated in different reading frames to yield distinct proteins (same locus, different product). One example in SARS-CoV-2, shown here, is the translation of protein ORF9b from an alternative (+1) reading frame of the N (nucleocapsid) gene. Protein structures show the N-terminal domain of N (Peng et al. 2020; Protein Data Bank 7CDZ) and the ORF9b homodimer (Weeks et al. 2020; Protein Data Bank 6Z4U), visualized using Mol* Viewer (Sehnal et al. 2021). The nucleotides shown correspond to coordinates 28282-98 in the reference genome Wuhan-Hu-1 (NC_045512.2), where N begins at 28274 and ORF9b begins at 28284 (for full coordinates of overlapping genes in SARS-CoV-2, see Jungreis, Nelson, et al. 2021).
Given the above, it is unfortunate that, to date, not a single out-of-frame gene has been annotated in the SARS-CoV-2 reference genome, Wuhan-Hu-1 (NC_045512.2). As a consequence, they are generally excluded from genomic, laboratory, and clinical analyses. Other frequently neglected accessory genes in SARS-CoV-2 include ORF6, ORF7a, ORF7b, ORF8, and the disputed ORF10.
Non-canonical genes are also documented in other pandemic viruses. This includes HIV-1, where the out-of-frame asp is expressed and integrated into the viral envelope (Affram et al. 2019) and is associated with pandemic spread (Cassan et al. 2016). Other examples come from such disparate viruses as influenza (Machkovech et al. 2019), betaherpesvirus (Finkel et al. 2020), and Zika virus (Irigoyen et al. 2017). One powerful approach for detecting such genes is ribosome profiling, which identifies actively translated mRNA fragments protected by ribosomes (i.e., ribosome footprints) (Stern-Ginossar 2015). Such new techniques for studying gene function provide opportunities for more inclusive studies of gene repertoire, particularly when characterizing newly emerged viruses.
Non-canonical genes demand a rethink of viral genome annotation and molecular biology. For example, requiring evolutionary conservation between virus lineages (Jungreis, Sealfon, and Kellis 2021) necessarily dismisses genes unique to one lineage (Nelson, Ardern, Goldberg, et al. 2020). Evolutionary and translatomic analyses of individual lineages (e.g., SARS-CoV-2 vs. SARS-CoV) together enable a more comprehensive understanding than standard methods based on codon usage, ORF length, or deep conservation (Nelson, Ardern, and Wei 2020). Indeed, non-canonical gene products interact with host cells and contribute to clinical outcomes, as demonstrated by ORF3b and ORF9b. We must stop neglecting non-canonical genes.
Acknowledgments
We thank Noam Stern-Ginossar for feedback on the text and Ming-Hsueh (Mitch) Lin for feedback on the figure.
References
Affram Y, Zapata JC, Gholizadeh Z, Tolbert WD, Zhou W, Iglesias-Ussel MD, Pazgier M, Ray K, Latinovic OS, Romerio F. 2019. The HIV-1 antisense protein ASP is a transmembrane protein of the cell surface and an integral protein of the viral envelope. J. Virol. 93:e00574-19. https://doi.org/10.1128/jvi.00574-19
Cassan E, Arigon-Chifolleau A-M, Mesnard J-M, Gross A, Gascuel O. 2016. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc. Natl. Acad. Sci. U. S. A. 113:11537–11542. https://doi.org/10.1073/pnas.1605739113
Finkel Y, Mizrahi O, Nachshon A, Weingarten-Gabbay S, Morgenstern D, Yahalom-Ronen Y, Tamir H, Achdout H, Stein D, Israeli O, et al. 2021. The coding capacity of SARS-CoV-2. Nature 589:125–130. https://doi.org/10.1038/s41586-020-2739-1
Finkel Y, Schmiedel D, Tai-Schmiedel J, Nachshon A, Winkler R, Dobesova M, Schwartz M, Mandelboim O, Stern-Ginossar N. 2020. Comprehensive annotations of human herpesvirus 6A and 6B genomes reveal novel and conserved genomic features. eLife 9:e50960. https://doi.org/10.7554/elife.50960
Firth AE. 2020. A putative new SARS-CoV protein, 3c, encoded in an ORF overlapping ORF3a. J. Gen. Virol. 101:1085–1089. https://doi.org/10.1099/jgv.0.001469
Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J. Gen. Virol. 93:1385–1409. https://doi.org/10.1099/vir.0.042499-0
Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, Tsang OTY, Yeung YC, Perera RAPM, Poon LLM, et al. 2020. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 21:1293–1301. https://doi.org/10.1038/s41590-020-0773-7
Ho JSY, Zhu Z, Marazzi I. 2021. Unconventional viral gene expression mechanisms as therapeutic targets. Nature 593:362–371. https://doi.org/10.1038/s41586-021-03511-5
Irigoyen N, Dinan AM, Meredith LW, Goodfellow I, Brierley I, Firth AE. 2017. The translational landscape of Zika virus during infection of mammalian and insect cells. bioRxiv. https://doi.org/10.1101/112904
Jungreis I, Nelson CW, Ardern Z, Finkel Y, Krogan NJ, Sato K, Ziebuhr J, Stern-Ginossar N, Pavesi A, Firth AE, et al. 2021. Conflicting and ambiguous names of overlapping ORFs in the SARS-CoV-2 genome: a homology-based resolution. Virology 558:145–151. https://doi.org/10.1016/j.virol.2021.02.013
Jungreis I, Sealfon R, Kellis M. 2021. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 12:2642. https://doi.org/10.1038/s41467-021-22905-7
Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ, USFQ-COVID19 Consortium, Nakagawa S, et al. 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 32:108185. https://doi.org/10.1016/j.celrep.2020.108185
Li Y, Xu Z, Lei Q, Lai D-Y, Hou H, Jiang H-W, Zheng Y-X, Wang X-N, Wu J, Ma M-L, et al. 2021. Antibody landscape against SARS-CoV-2 reveals significant differences between non-structural/accessory and structural proteins. Cell Rep. 36:109391. https://doi.org/10.1016/j.celrep.2021.109391
Machkovech HM, Bloom JD, Subramaniam AR. 2019. Comprehensive profiling of translation initiation in influenza virus infected cells. PLoS Pathog. 15:e1007518. https://doi.org/10.1371/journal.ppat.1007518
Nagler A, Kalaora S, Barbolin C, Gangaev A, Ketelaars SLC, Alon M, Pai J, Benedek G, Yahalom-Ronen Y, Erez N, et al. 2021. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA-peptidomics. Cell Rep. 35:109305. https://doi.org/10.1016/j.celrep.2021.109305
Nelson CW, Ardern Z, Goldberg TL, Meng C, Kuo C-H, Ludwig C, Kolokotronis S-O, Wei X. 2020. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. eLife 9:e59633. https://doi.org/10.7554/elife.59633
Nelson CW, Ardern Z, Wei X. 2020. OLGenie: Estimating natural selection to predict functional overlapping genes. Mol. Biol. Evol. 37:2440–2449. https://doi.org/10.1093/molbev/msaa087
Pavesi A. 2021. Origin, evolution and stability of overlapping genes in viruses: a systematic review. Genes 12:809. https://doi.org/10.3390/genes12060809
Peng Y, Du N, Lei Y, Dorje S, Qi J, Luo T, Gao GF, Song H. 2020. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 39:e105938. https://doi.org/10.15252/embj.2020105938
Sehnal D, Bittrich S, Deshpande M, Svobodová R, Berka K, Bazgier V, Velankar S, Burley SK, Koča J, Rose AS. 2021. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 49:W431–W437. https://doi.org/10.1093/nar/gkab314
Stern-Ginossar N. 2015. Decoding viral infection by ribosome profiling. J. Virol. 89:6164–6166. https://doi.org/10.1128/JVI.02528-14
Thorne LG, Bouhaddou M, Reuschl A-K, Zuliani-Alvarez L, Polacco B, Pelin A, Batra J, Whelan MVX, Ummadi M, Rojc A, et al. 2021. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. https://doi.org/10.1101/2021.06.06.446826
Weeks SD, De Graef S, Munawar A. 2020. X-ray crystallographic structure of Orf9b from SARS-CoV-2. https://www.wwpdb.org/pdb?id=pdb_00006z4u
Weingarten-Gabbay S, Klaeger S, Sarkizova S, Pearlman LR, Chen D-Y, Gallagher KME, Bauer MR, Taylor HB, Dunn WA, Tarr C, et al. 2021. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell 184:3962-3980.e17. https://doi.org/10.1016/j.cell.2021.05.046
Zecha J, Lee C-Y, Bayer FP, Meng C, Grass V, Zerweck J, Schnatbaum K, Michler T, Pichlmair A, Ludwig C, et al. 2020. Data, reagents, assays and merits of proteomics for SARS-CoV-2 research and testing. Mol. Cell. Proteomics 19:1503–1522. https://doi.org/10.1074/mcp.RA120.002164