Posted on behalf of PFHVG
National authorities in Guinea declared a new Ebolavirus (EBOV) disease outbreak in Nzérékoré region on 14 February 2021. To identify the EBOV strain responsible for this outbreak, five samples were sequenced at PFHVG using MinION (Oxford Nanopore) according to the ARTIC network protocol (https://artic.network). A consensus ranging from 47% to 88% was obtained for 4 samples while only 7.5% consensus genome was recovered from one sample (not included in the analyses below).
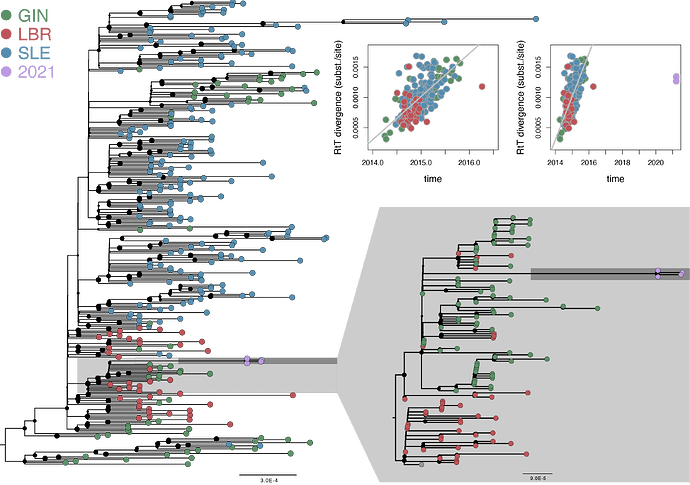
The 4 sequences were identified as the Makona variant (Zaire ebolavirus) that was responsible for the 2013-2016 EBOV outbreak in West Africa. Phylogenetic reconstruction demonstrates that the four sequences cluster together within the Makona diversity (Fig. 1), indicating that the current outbreak is the result of human transmission rather than an new introduction from the animal reservoir. Remarkably, the branch ancestral to this cluster represents only 12 substitutions, much less than would be expected for genomes sampled in 2021 based on the EBOV evolutionary rate during the 2013-2016 EBOV outbreak (Fig. 1). This indicates a long period of latency (5 to 7 years) prior to the this year’s resurgence. This extends previous findings of a flare-up in 2016 in Guinea originating from a survivor with virus persistence in seminal fluid for more than 500 days (Diallo et al., 2016).
Fig. 1. Maximum likelihood phylogenetic reconstructions based on a data set of 300 Makona strains and four 2021 genomes. The dark grey box highlights the cluster for which a separate tree was inferred based on 119 Makona strains. The dark grey box highlights the new 2021 lineage. The plot insets show root-to-tip divergence as a function of sampling time for the 300 Makona strains (left panel, the outlier point represents a genome that derives from the 2016 flare-up) and for the same strains with the 4 2021 genomes (right panel).
April 20 update
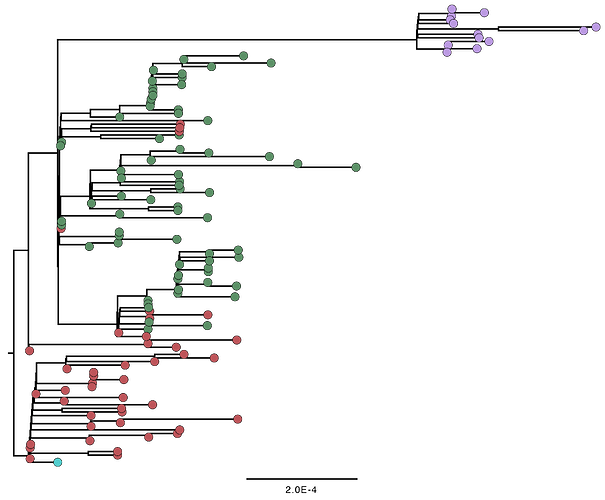
9 additional genomes confirm the pattern described above.
Fig. 2. Maximum likelihood phylogenetic reconstruction for the same cluster as analysed in the grey inset in Fig. 1.
The genome data is available here: GitHub - PFHVG/EBOVsequencing: repository for Ebola virus sequencing at PFHG in Guinea, 2021
DISCLAIMER
The unpublished sequences are shared via the website virological.org under the following conditions: The Ebola virus sequences from Guinea are shared by the Laboratoire du “Projet des Fièvres Hémorragiques Virales de Guinée” (PFHVG), Conakry, Guinée, to support the public health response as well as the development and evaluation of Ebola virus disease diagnostics and therapeutics. The data may be used and analyzed for these purposes. We are preparing a scientific publication of the data. In the meantime, this post may be cited. If you intend to use the sequences for publication prior to our own publication, please contact us directly.
Dr. Magassouba N’Faly < [email protected]>, Director of the Laboratory “Projet des Fièvres Hémorragiques de Guinée” (PFHG), Conakry, Guinea.
The sequencing work at the Laboratoire du PFHGV is supported by WHO GOARN, the Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany, and other technical partners.
Technical partners are represented by Prof. Stephan Günther [email protected], WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Reference and Research at BNITM.
References
Diallo B, Sissoko D, Loman NJ, et al. Resurgence of Ebola Virus Disease in Guinea Linked to a Survivor With Virus Persistence in Seminal Fluid for More Than 500 Days. Clin Infect Dis. 2016;63(10):1353-1356. doi:10.1093/cid/ciw601