Mariana Soares da Silva¹, Juliana Schons Gularte¹, Ana Cristina Sbaraini Mosena¹, Meriane Demoliner¹, Micheli Filippi¹, Alana Witt Hansen¹, Vyctoria Malayhka de Abreu Góes Pereira¹, Fágner Henrique Heldt¹, Matheus Nunes Weber¹, Paula Rodrigues de Almeida¹, Juliane Deise Fleck¹, Fernando Rosado Spilki¹
¹Universidade Feevale, Laboratório de Microbiologia Molecular, Rodovia ERS-239, Nº
2755, Prédio Vermelho, Piso 1, sala 103, Vila Nova, CEP 93525-075, Novo Hamburgo,
RS, Brazil.
In November 2021, researchers in Botswana and South Africa identified a new SARS-CoV-2 variant through whole-genome sequencing (WGS), posteriorly named as Omicron (B.1.1.529) [1]. This variant, announced by WHO as a Variant of Concern (on November 26, 2021), presents more than 50 mutations in its genome; of which, 32 were in the receptor binding domain (RBD) of the spike protein [2]. Omicron has spread rapidly, increasing COVID-19 cases through the world [3]. In Brazil, the first description was reported a few weeks after the first case in South Africa, from an airplane passenger that arrived in São Paulo State from South Africa, in late November.
A total of 87 SARS-CoV-2 complete genomes were sequenced through Ilumina MiSeq platform at Laboratório de Microbiologia Molecular - Feevale University, one of the institutions linked to the Corona-ômica.BR-MCTI Network. The positive samples were collected between late November and mid-January, from patients belonging to eleven cities from Rio Grande do Sul State (Arroio do Meio (1), Campo Bom (15), Canoas (9), Estância Velha (1), Garibaldi (5), Ivoti (1), Novo Hamburgo (41), Santa Cruz do Sul (11), São Sebastião do Caí (1), Sobradinho (1) and Triunfo (1).
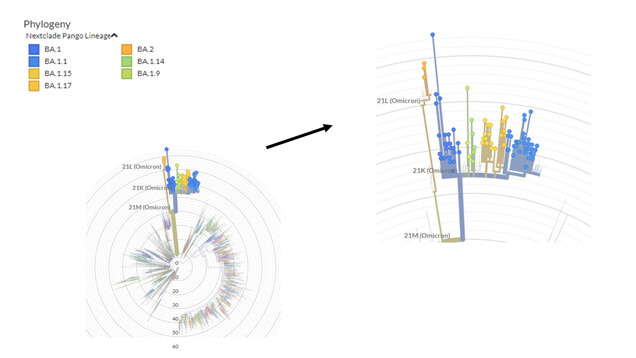
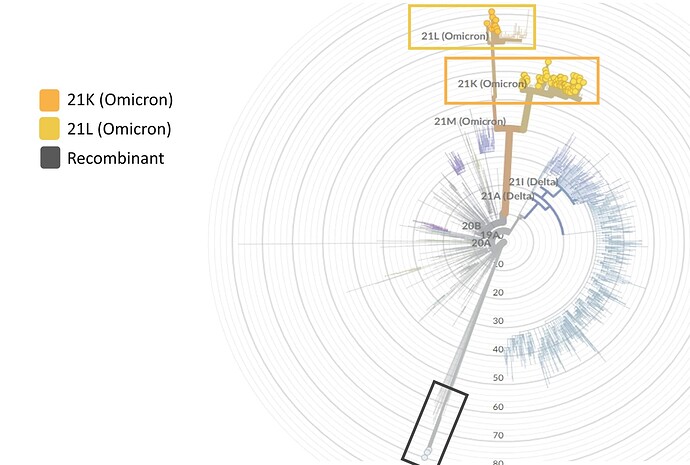
Regarding genetic characterization, the sequences were aligned with complete SARS-CoV-2 genomes of different lineages through the NextClade online tool (https://clades.nextstrain.org/) and the phylogenetic tree was inferred (Figure 1). Of the total samples, 26 (30%) were classified as Delta variant (21J) and 61 (70%) as Omicron (21K). Subsequently, the sequences were analyzed through the Pangolin online tool (https://github.com/hCoV-2019/pangolin) and characterized as belonging to the following Delta: AY.99.2 (15/26 – 57.7%), AY.101 (3/26 – 11.5%), AY.122 (2/26 – 7.6%), AY.43 (2/26 – 7.6%), AY.112 (1/26 – 3.9%), AY.25 (1/26 – 3.9%), AY.43.2 (1/26 – 3.9%), AY.46 (1/26 – 3.9%) and Omicron sub lineages: BA.1 (38/61 – 62%) and BA.1.1 (23/61 – 38%).
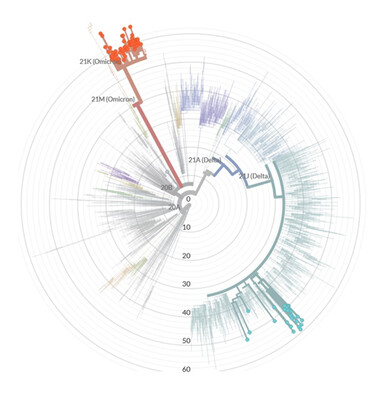
Figure 1. Phylogenetic tree performed through NextClade online tool. In green, 26 sequences characterized as Delta variant and in red 61 Omicron variant sequences.
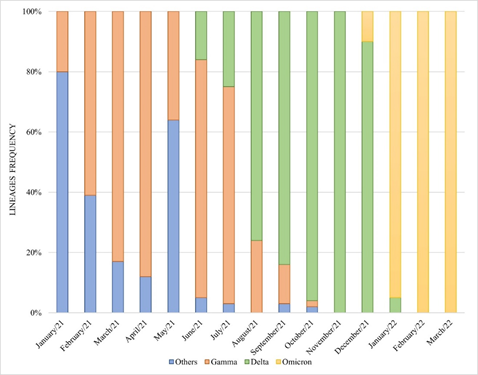
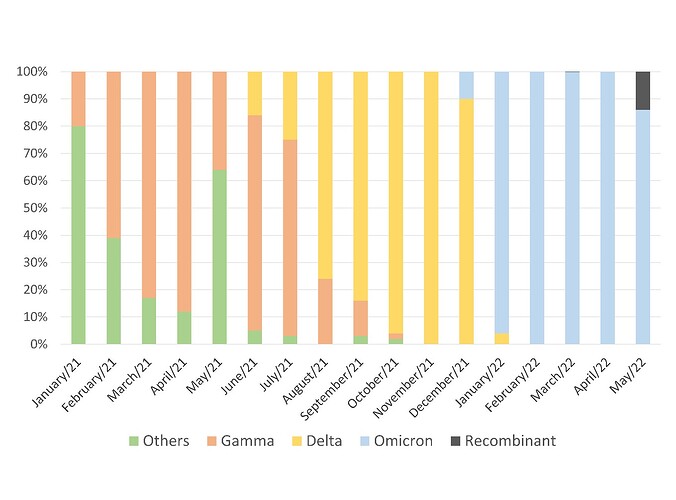
Performing a retrospective analysis, it is interesting to note that, among the samples sequenced herein, the Delta variant represented 100% of frequency in November 2021. In December of the same year, the first Omicron variant detections were performed, however not very expressive (3/30 – 10%), and the Delta variant still predominated (27/30 – 90%). Less than a month later, in January 2022, the scenario reversed, and the Delta comprised only 4 samples (4/62 – 6%) while Omicron established with 58 detections (58/62 – 94%). The complete data can be seen in Figure 2.
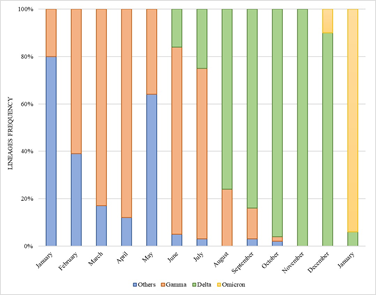
Figure 2. Lineages distribution of the SARS-CoV-2 samples sequenced by Laboratório de Microbiologia Molecular at Feevale University between January 2021 and January 2022.
According to our results, Omicron variant established in a short period of time, which was also observed in other countries [3]. It spreads faster and can evade natural and vaccine-induced immunity better than its predecessor variants [4]. Even the milder severity, especially in fully vaccinated individuals, the public health impact should be strongly considerable since the cases increased exponentially. It is likely that others SARS-CoV-2 variants will emerge and the continuous case control as well as epidemiological genomic surveillance must continue.
References
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. Geneva, Switzerland: World Health Organization; 2021. Accessed December 3, 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern
- Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21.
- Mohapatra, R.K., Sarangi, A.K., Kandi, V., Azam, M., Tiwari, R. and Dhama, K. (2022), Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: Current global scenario. J Med Virol. https://doi.org/10.1002/jmv.27561.
- Rössler, A., Riepler, L., Bante, D., Laer, D. von & Kimpel, J. SARS-CoV-2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv (2021) https://doi.org/10.1101/2021.12.08.21267491.