Potential novel SARS-CoV-2 variant of interest from lineage AP.1 with N501Y and additional spike mutations identified in Saxony, Germany
Severinov DA1, Yi B3, Dalpke A3, Rost F4,5, Winkler S6, Beil J6, Reinhardt S4, Klemroth S4, Mehnert G4, Hartkopf F9, Hölzer M7, Kühnert D8, German COVID-19 OMICS Initiative (DeCOI)10, Poetsch AR1,2 *.
1Biomedical Genomics, Biotechnology Center, TU Dresden, Germany
2Biomedical Genomics, NCT Dresden, Germany
3Institute of Medical Microbiology and Virology, TU Dresden, Germany
4Dresden Concept Genome Center, TU Dresden, Germany
5Center for Regenerative Therapies Dresden, TU Dresden, Germany
6MPI-CBG, Dresden and Dresden Concept Genome Center, TU Dresden, Germany
7Methodology and Research Infrastructure, MF1 Bioinformatics, Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany
8Transmission, Infection, Diversification & Evolution Group, Max-Planck Institute for the Science of Human History; Kahlaische Str. 10, 07745 Jena, Germany.
9Methodology and Research Infrastructure, MF2 Bioinformatics, Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany
*Correspondence: [email protected]
Since the onset of the pandemic triggered by SARS-CoV-2, the virus has accumulated a large number of additional mutations. Most mutations do not alter the transmissibility and infectivity of the virus considerably, if at all. However, a few, particularly in the spike gene, have been shown to increase the pathogenicity of the virus. Therefore, routine molecular surveillance has been implemented in several countries, including Germany, and efforts have been boosted in Germany since February 2021, to monitor SARS-CoV-2 evolution closely.
Novel strains with the spike 501Y variant currently outcompete previous strains with spike 501N. They have evolved convergently in several places around the world 1-4. Some have been described as more infectious, leading to a more severe disease. Furthermore, the accumulation of additional mutations has shown to even overcome immunity acquired through infection with earlier variants 2.
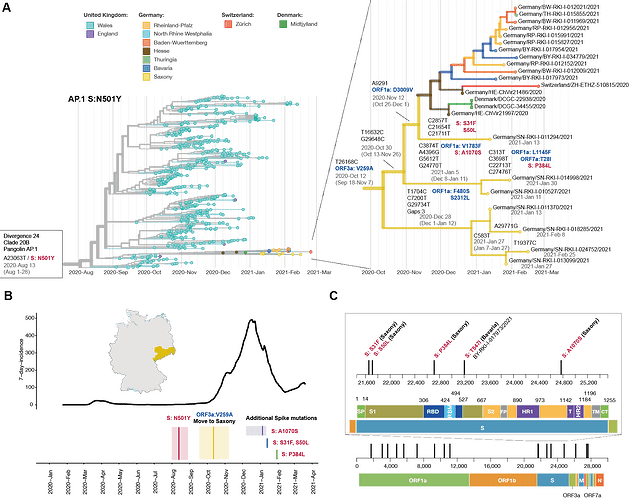
The spike N501Y mutation has also evolved in less common lineages, where it tends to persist, leading to additional lineages that accumulate additional mutations with a spike N501Y background. One such lineage is AP.1 (https://cov-lineages.org/lineages/lineage_AP.1.html) 5, the “Wales lineage” descending from B.1.1.70 within clade 20B. With already 23 mutations difference to the Wuhan reference virus, the lineage additionally evolved the spike mutation N501Y and was subsequently predominantly detected in Wales (Fig.1A). The mutation is estimated to have emerged around Aug 13, 2020 (Aug1-28). Due to uneven sequencing efforts among different countries, it is difficult to assess when and where the lineage has spread. However, mobility of the virus to Saxony, Germany, can be detected, coinciding with an additional mutation in ORF3a: V259A. This mutation, and consequently the time frame of mobility can be estimated to have occurred around Oct 12, 2020 (Sep 18 - Nov 7). At this time, incidences of SARS-CoV-2 cases in Saxony entered an exponential growth phase and the lineage had the possibility to spread and accumulate additional mutations. Seven viruses could be detected in Saxony to this date with a total addition of 18 mutations. Ten of these are non-synonymous protein-coding, five in ORF1a, one in ORF7a, and four in the spike proteins; S31F, S50L, P384L, A1070S (Fig. 1C). The spike protein mutations evolved in the timeframe before the end of Feb 2021, when incidence numbers were particularly high in Saxony (Fig. 1B).
In parallel, the lineage also spread to other parts of Germany, Denmark and Switzerland. One branch accumulated an additional mutation in spike (T547I), found in Bavaria.
Mutations in the spike protein are of particular concern when they occur in the N-terminal domain (NTD), particularly in or near the receptor-binding domain (RBD) or the receptor binding motif (RBM), the domains that interact with the ACE2 receptor. Three of the variants fulfill this description. S31F and S50L are located in the NTD, yet not in the antibody binding domain. As of 29 March 2021 they have been seen to evolve together in lineage B.1.5.96 in the USA (outbreak.info), and moreover have occurred independently before. At the same time, S31F has been detected in 17 viruses in several countries and lineages, likely to have evolved convergently. S50L has been detected in 123 viruses in several countries and lineages. Interestingly, the occurrence of both mutations together exceeds expectations (p<10-5, hypergeometric test). The third mutation, P384L lies in the RBD. This mutation has been detected in 799 viruses in 35 countries and multiple lineages, yet is still present in less than 0.5 % of cases worldwide. Similarly, at the time of writing, there is insufficient evidence to raise serious concern for the other spike mutations.
However, this local example shows that high incidence numbers provide an evolutionary opportunity for additional mutations on top of the mutation of concern, spike N501Y. High incidence numbers increase the chances for adaptation of the virus and convergent evolution with additional mutations followed by successful propagation. It also highlights that AP.1 should be carefully monitored, as should every variant that convergently evolves the spike N501Y mutation. A focussed mutation screening of the area where it is most prevalent would allow investigating how it competes against other lineages. This would also enable early detection, should any of the additional mutations be of concern or should the virus develop additional mutations that may become of concern.
Figure 1: Lineage AP.1. accumulates additional spike mutations in Saxony Germany. (A) phylogenetic tree representation of the AP.1. lineage with a spike N501Y mutation. The branch after migration to Saxony is highlighted with a focus on the nucleotide changes and their coding effect for the viruses detected in Saxony. Time estimates are derived from the phylogenetic tree, depicted with the confidence interval. Virus IDs correspond to the identifiers in the GISAID database. (B) 7-day-incidence statistics per 100.000 inhabitants for Saxony highlighting the timepoints of spike mutations and the virus migration from Wales to Saxony. Time confidence intervals are derived from the branches of the phylogenetic tree, unless the mutation is only represented by one virus, in which case the sampling time is shown. (C) Coding mutations and their locations in the virus genome for all viruses following the migration to Saxony. Zoom into the Spike protein (S) to highlight the associated mutations and their distribution within the spike domains; S1 = SARS-CoV-2 attachment subunit; S2 = SARS-CoV-2 fusion subunit; SP = signal peptide; RBD = receptor binding domain; RBM = receptor binding motif; FP = fusion peptide; HR1 = heptad repeat 1 region; HR2 = heptad repeat 2 region; T = tether region; TM = transmembrane domain; CT = cytoplasmic tail.
Methods:
Data were obtained on 29 March 2021 from GISAID 6,7. 7-day-incidence statistics were obtained for the different regions from Saxony from the Robert Koch Institute (https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Daten/Fallzahlen_Archiv.html), combined and normalised to the total population of Saxony and visualised with R. Genome annotation was obtained from nextstrain.org 8, and further differentiated for the spike protein 9. Phylogenetics were performed with nextstrain 8, using global data with a subsample focus on Saxony and fixed inclusion of the pangolin AP.1 lineage. Overlap statistics of co-occurrence of mutations was performed with a hypergeometric test (http://nemates.org/MA/progs/overlap_stats.html) conservatively assuming a total population of 500.000 viruses. Given that the individual occurrences of the mutations are not independent, yet the co-occurrence has evolved independently, the resulting level of significance is an underestimate.
Acknowledgments:
We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based. We specifically thank all staff from the laboratories in Saxony for their sequencing efforts, including the Dresden Concept Genome Center. We thank the Robert Koch Institute for the data management and submission to GISAID and the German and Saxonian Ministries of Health for funding the sequencing efforts. We thank the team of nextstrain.org for providing their data analysis platform, making it accessible, and providing superb support. We thank DeCOI for the continous network effort in supporting OMICS research on COVID-19 in Germany. Anna Poetsch receives funding from the Mildred Scheel Early Career Center Dresden funded by the German Cancer Aid.
German COVID-19 OMICS Initiative (DeCOI):
Janine Altmüller, Angel Angelov, Robert Bals, Alexander Bartholomäus, Anke Becker, Daniela Bezdan, Ezio Bonifacio, Peer Bork, Nicolas Casadei, Thomas Clavel, Maria Colome-Tatche, Andreas Diefenbach, Alexander Dilthey, Nicole Fischer, Konrad Förstner, Sören Franzenburg, Julia-Stefanie Frick, Gisela Gabernet, Julien Gagneur, Tina Ganzenmüller, Marie Gauder, Alexander Goesmann, Siri Göpel, Adam Grundhoff, Torsten Hain, André Heimbach, Michael Hummel, Thomas Iftner, Angelika Iftner, Stefan Janssen, Jörn Kalinowski, René Kallies, Birte Kehr, Andreas Keller, Sarah Kim-Hellmuth, Christoph Klein, Oliver Kohlbacher, Karl Köhrer, Jan Korbel, Peter G. Kremsner, Denise Kühnert, Ingo Kurth, Markus Landthaler, Yang Li, Kerstin Ludwig, Oliwia Makarewicz, Manja Marz, Alice McHardy, Christian Mertes, Sven Nahnsen, Markus Nöthen, Francine Ntoumi, Peter Nürnberg, Stephan Ossowski, Jörg Overmann, Silke Peter, Klaus Pfeffer, Anna R. Poetsch, Alfred Pühler, Nikolaus Rajewsky, Markus Ralser, Olaf Rieß, Stephan Ripke, Ulisses Nunes da Rocha, Philip Rosenstiel, Antoine-Emmanuel Saliba, Leif Erik Sander, Birgit Sawitzki, Philipp Schiffer, Wulf Schneider, Eva-Christina Schulte, Joachim L. Schultze, Alexander Sczyrba, Yogesh Singh, Michael Sonnabend, Oliver Stegle, Jens Stoye, Fabian Theis, Janne Vehreschild, Thirumalaisamy P. Velavan, Jörg Vogel, Max von Kleist, Andreas Walker, Jörn Walter, Dagmar Wieczorek, Sylke Winkler, John Ziebuhr
References:
- Rambaut, A. et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org (2020).
- Faria, N. R. et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological.org (2021).
- Martin, D. P. et al. The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape. medRxiv (2021) doi:10.1101/2021.02.23.21252268.
- Tegally, H. et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv (2020) doi:10.1101/2020.12.21.20248640.
- Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5, 1403–1407 (2020).
- Elbe, S. & Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 1, 33–46 (2017).
- Shu, Y. & McCauley, J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 22, (2017).
- Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123 (2018).
- Lan, J. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020).