Like most of you I (Sam Lycett), and my undergraduate project student (Rhys Inward) have also been processing the GISAID data, especially now that there are ‘local’ sequences from Glasgow and Edinburgh (many many thanks to all those involved especially Andrew Rambaut and team, and Dave Robertson and team).
Phylogeography
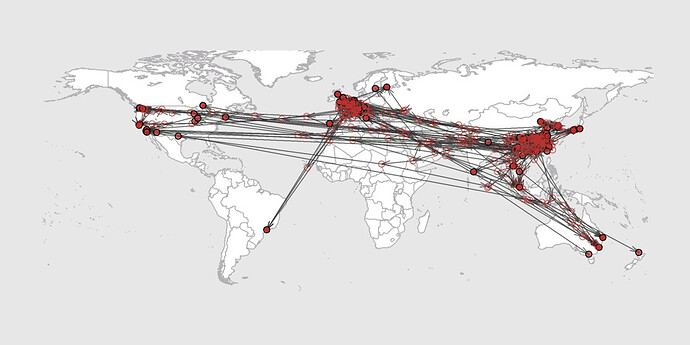
Here is our phylogeography with data until 11 March (we’re still working on the data until 21 March).
Data: 247 whole genome GISAID sequences (removed the ones with too many N’s but kept rather more Edinburgh and Glasgow); aligned with MAFFT, manual adjust, trim to start of ORF1ab, truncate to end of last ORF (so no 5’UTR or 3’UTR, but this is not codon aligned – we didn’t put the frameshifts in for the phylogeography).
Model: BEAST 1.10.4 time-scaled trees - TN93 + 4 gamma categories, strict clock with lognormal prior (real space mean = 1e-3, std = 1e-3), exponential growth tree / population prior. Then re-use 1000 from the posterior sample post burn-in as empirical trees and apply homogeneous brownian motion with lat-lon & jitter = 0.001 (lat-lon data to nearest city where possible, manual look up in some cases)
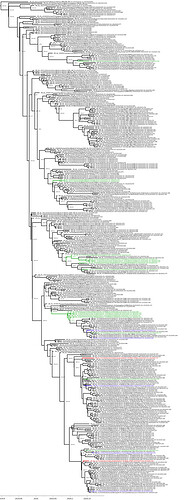
BEAST with UK sequences
For the latest on the Scottish sequences see the excellent posts by Dave Robertson and team
Data: 373 whole genome GISAID sequences (removed the ones with too many N’s but kept rather more Edinburgh and Glasgow); aligned with MAFFT, manual adjust, trim to start of ORF1ab, truncate to end of last ORF. There are 5 sequences from Scotland, 17 from England and 2 from Wales in this alignment.
Model: Model: BEAST 1.10.4 time-scaled trees - TN93 + 4 gamma categories, strict clock with lognormal prior (real space mean = 1e-3, std = 1e-3), exponential growth tree / population prior.
England = Green, Scotland = Blue, Wales = Red
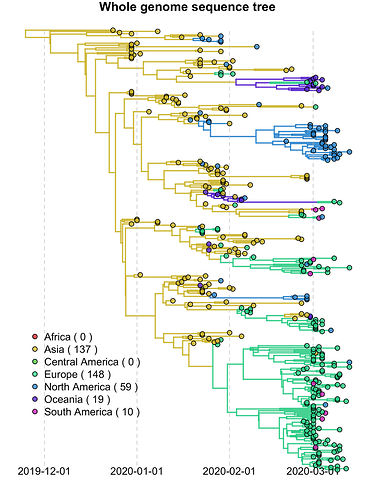
BEAST by Continent
Data and model as above (373 genomes), but with Continent added as discrete trait when using 1000 empirical trees (symmetric model with BSSVS). Here you can more easily see the separate introductions and subsequent spread into Europe, North America and Oceania.
Discussion points and further analysis
I think there might now be enough time depth and variation to try the Birth Death Multi-Type tree on just Asia and Europe, to try and get at the individual growth rates (am running this now). And looking beyond this, hopefully we (all) will be in a position soon to try and estimate whether the social distancing interventions are indeed having an impact on the inferred epidemiological parameters (epoch models).
Data Acknowledgements
Huge thanks to the efforts of all who are submitting to GISAID - your work is vital ! Below is the acknowledgment table for all the strains with the submitting lab details, and also the table of the subset that I used together with processed location details.
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu™ Database on which this research is based.
gisaid_cov2020_acknowledgement_table.xls.zip (77.5 KB)
COVID-19_human_373_b_info.txt.zip (11.8 KB)