Multiplexed RT-qPCR to screen for SARS-COV-2 B.1.1.7 variants: Preliminary results
Chantal B.F. Vogels, Tara Alpert, Mallery Breban, Joseph R. Fauver, Nathan D. Grubaugh
Yale School of Public Health
Contact: [email protected]
Tl;dr - we developed an open-source multiplexed PCR that recreates the TaqPath SGTF signature. Protocol here. We are further optimizing it to differentiate between B.1.1.7 samples and other variants.
Introduction
As the B.1.1.7 lineage "UK" variant continues to spread, there is an urgent need for labs to rapidly screen COVID-19 samples for these viruses. While sequencing is the gold standard, it cannot always be scaled or implemented in some settings. The Applied Biosystems TaqPath COVID-19 PCR assay (ThermoFisher) was discovered to have a distinct signature (spike gene target failure, [SGTF]) when testing viruses containing the Δ69/70 HV deletion, including the B.1.1.7 variant. Detecting the Δ69/70 HV deletion alone is not definitive for the B.1.1.7 variant as this deletion has arisen independently in other lineages, but tracking the frequencies of SGTFs helped the UK to track the B.1.1.7 variant. In the US and other countries, screening samples for the SGTF helped to identify potential B.1.1.7 variants for sequencing prioritization. ThermoFisher, however, has not released their spike probe sequence, so the assay needs to be recreated to be used more broadly.To differentiate between B.1.1.7 and other variants, we designed primers and probes to the Δ69/70 HV deletion and the Δ144 Y deletion. We posted the draft protocol on Twitter on 31 Dec 2020. Here we provide the first results of that assay. While the Δ144 Y set requires further optimization, the Δ69/70 HV set was able to distinguish between variant and non-variant samples similar to the TaqPath SGTF signature. We combined the Δ69/70 HV set in a multiplexed PCR assay with the CDC N1 set as a positive control, and the CDC RNase P set as an extraction/sample control as an open source method to screen for viruses like B.1.1.7 with the Δ69/70 HV deletion. Essentially it is an open-source “hack” of the TaqPath assay.
Methods
Ethics
PCR testing and sequencing of de-identified remnant COVID-19 clinical samples was conducted in accordance with the guidelines of the Yale Human Investigations Committee.Specimens
From our routine sequencing program, we have selected 8 SARS-CoV-2 samples belonging to a variety of strains without the Δ69/70 HV and Δ144 Y deletions, as well as 6 SARS-CoV-2 samples belonging to the B.1.375 lineage (Δ69/70 HV deletion), and 2 SARS-CoV-2 samples belonging to the B.1.1.7 lineage (Δ69/70 HV and Δ144 Y deletions). All samples were nasal or nasopharyngeal swabs, and nucleic acid was extracted from 300 µL viral transport medium and eluted in 75 µL using the MagMAX viral/pathogen nucleic acid isolation kit (ThermoFisher Scientific, Waltham, MA).Primer and probe design
We developed primer/probe sets targeting the Δ69/70 HV and Δ144 Y deletions to create an open-source RT-qPCR protocol for B.1.1.7 screening, because the spike probe sequence as used in the TaqPath COVID-19 combo kit is not publicly available. We aligned the SARS-CoV-2 reference genome with genomes from B.1.1.7 viruses to design primers flanking the Δ69/70 HV and Δ144 Y deletions, and probes that cover both deletions. For each deletion we designed two probes either targeting the reference virus (drop-out) or the B.1.1.7 virus (detection). For the “drop-out” probes we were expecting to see no amplification with B.1.1.7 lineage viruses and amplification with other viruses, whereas for the “detection” assay we were expecting the opposite with amplification of B.1.1.7 viruses and no amplification of other viruses.RT-qPCR conditions
In our preliminary evaluation, we tested both the “detection” and “drop-out” assay which included the CDC N1 primer/probe as a SARS-CoV-2 detection control (Lu et al. 2020), as well as the two primer/probe sets targeting the Δ69/70 HV and Δ144 Y deletions in the spike gene (**Table 1**). We followed the same conditions as described in our multiplex RT-qPCR assay as previously described (Kudo et al. 2020). In brief, we used the Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs, Ipswich, MA) with 400 nM of each primer and 200 nM of each probe per reaction, and 5 µL of RNA in a total reaction volume of 20 µL. We ran separate assays using either the 69/70del and 144del detection or drop-out probes. PCR cycler conditions were reverse transcription for 10 min at 55 °C, initial denaturation for 1 min at 95 °C, followed by 45 cycles of 10 s at 95 °C and 30 s at 55 °C on the Biorad CFX96 touch real-time PCR detection system (Bio-Rad, Hercules, CA). Based on our preliminary findings we have selected the primer/probe sets that worked best and published the detailed protocol online (Vogels, Fauver, and Grubaugh 2021). - Results
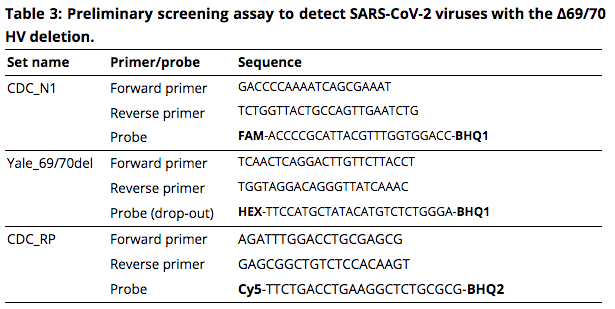
We aimed to develop a RT-qPCR assay to facilitate the screening for the rapidly emerging B.1.17 lineage viruses. By testing a selection of SARS-CoV-2 samples with and without the Δ69/70 HV and Δ144 Y deletions, we found that our Yale_69/70del “drop-out” primer/probe set can serve as a screening tool for detection of viruses with the Δ69/70 HV deletion (**Table 2**). The Yale_69/70del “detection” primer/probe set resulted in detection of all, except 2, samples, and is therefore not suitable for detection of Δ69/70 HV variants. We also included a Yale_144del primer/probe set for detection or drop-out of the 3 nucleotide Δ144 Y deletion, which could have helped to differentiate between B.1.1.7 viruses and other viruses such as B.1.375 which have the Δ69/70 HV deletion. However, we found detection of almost all samples with both the detection and drop-out set, and therefore the design of the Yale_144del primer/probe sets needs to be further optimized to meet its goal. Thus, the Yale_69/70del (drop-out) primer/probe set can be used for screening of SARS-CoV-2 variants with the Δ69/70 HV deletion, and further optimization is needed to design an assay that can specifically differentiate between B.1.1.7 and other viruses that contain the 6 nucleotide deletion. - Based on our preliminary findings, we propose a Δ69/70 HV deletion RT-qPCR screening assay that can help to prioritize samples for sequencing in areas where the B.1.1.7 lineage viruses may circulate at low frequency. This screening assay is for research purposes only, and cannot be used for clinical diagnosis. The assay consists of the CDC N1 and human RNase P (RP) primer/probe sets (Lu et al. 2020), which serve as virus and human controls, respectively, in combination with the newly designed Yale_69/70del (drop-out) primer/probe set (Table 3). A detailed protocol has been made openly available immediately after the initial validation (Vogels, Fauver, and Grubaugh 2021).
Discussion
The rapid emergence of the SARS-CoV-2 lineage B.1.1.7 requires the need for increased screening of this highly transmissible virus. Although sequencing is the golden standard in detection of new variants, labor and cost limit the number of samples that can be sequenced. We designed a RT-qPCR B.1.1.7 screening assay to help prioritize samples for sequencing. We aimed to develop an assay that can differentiate between B.1.1.7 viruses (Δ69/70 HV and Δ144 Y deletions) and other viruses such as B.1.375 that have the Δ69/70 HV deletion as well. Our preliminary validations show that our RT-qPCR assay can detect viruses with the Δ69/70 HV deletion by drop-out of the Yale-69/70del primer/probe set, which helps to prioritize samples for sequencing.Our RT-qPCR assay is still under development and we wanted to post our preliminary findings online to make this information available to other labs as soon as possible. Given the preliminary stage of development, there are some important limitations to the study as presented here. First, we only tested a very low number of samples and are continuing to test more samples (with and without the Δ69/70 HV and Δ144 Y deletions) to increase our samples size. Second, the current assay is able to identify so-called spike gene target failure samples and can serve as a strategy to prioritize samples for sequencing, but it does not specifically detect B.1.1.7 lineage viruses. We are further optimizing the Yale_144del primer/probe set to enable differentiation of B.1.1.7 viruses from other viruses with the Δ69/70 HV deletion which has independently arisen in other lineages such as the newly identified B.1.375 lineage. Third, additional validation is needed to determine the sensitivity of the current assay, by testing samples with higher cycle threshold (Ct) values which could lead to false drop-out. In conclusion, our assay can serve as a screening assay to facilitate the selection of high priority samples for sequencing to detect the emerging B.1.1.7 lineage, but it should not be used as a definitive assay for B.1.1.7 detection.
References
Kudo, Eriko, Benjamin Israelow, Chantal B. F. Vogels, Peiwen Lu, Anne L. Wyllie, Maria Tokuyama, Arvind Venkataraman, et al. 2020. “Detection of SARS-CoV-2 RNA by Multiplex RT-qPCR.” bioRxiv. https://doi.org/10.1101/2020.06.16.155887. - Lu, Xiaoyan, Lijuan Wang, Senthilkumar K. Sakthivel, Brett Whitaker, Janna Murray, Shifaq Kamili, Brian Lynch, et al. 2020. “US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2.” Emerging Infectious Diseases 26 (8). https://doi.org/10.3201/eid2608.201246.Vogels, C. B. F., J. R. Fauver, and N. D. Grubaugh. 2021. “Multiplexed RT-qPCR to Screen for SARS-COV-2 B.1.1.7 Variants.” January 12, 2021. Multiplexed RT-qPCR to screen for SARS-COV-2 B.1.1.7 variants protocol materials.