Erroneous Mutations Associated with 64_L-60_R Primer-Dimer in ARTIC 4/4.1
Report prepared by: Sam Wilkinson, Natalie Groves, Josh Quick, Nick Loman
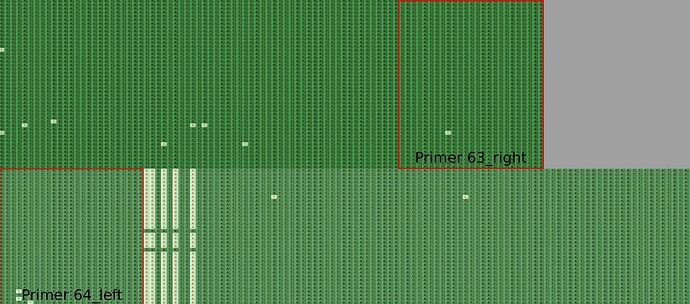
Recently 664 (correct as of 2022-03-29) SARS-CoV-2 sequences have been detected within the COG-UK dataset with the following mutations; T19209G, A19210G, A19212G, A19214G, C19217A located near the start ofARTIC V4/4.1 amplicon 64 (Figure 1). However these mutations were i) never present within overlapping reads in amplicon 63, ii) were not present in all reads in amplicon 64, suggesting that these mutations are artifactual.
Figure 1
The “GGGGTGTCA’’ motif is present a single time in the SARS-CoV-2 reference genome at positions 18,315-18,323. Notably this is a region covered by the 3’ region of ARTIC V4/4.1 amplicon 60. We determined the potential for interactions between primer 60_RIGHT and 64_LEFT and found a cross-primer dimerisation (Figure 2 and 3).
Figure 2
Figure 3
These primers contribute to amplicons that are generally underrepresented in sequencing, suggesting that primer-dimer formation may be occurring. This formation is likely to consume the available 64_LEFT during PCR after which the dimer product begins to prime the reaction, leading to the incorporation of the primer-dimer into the sequencing results and the artifactual variant calls observed.
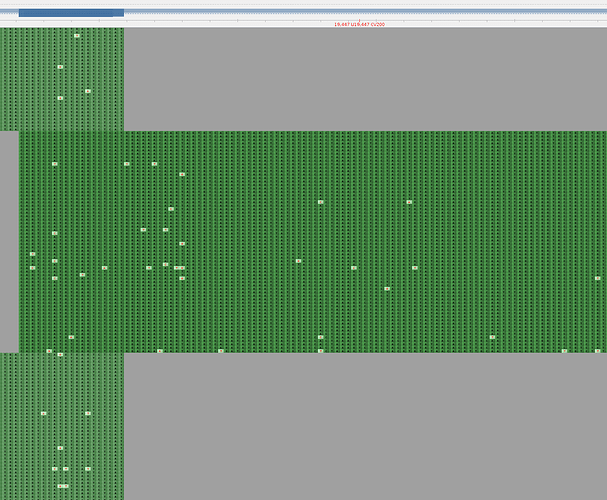
One possible solution to filter these erroneous calls is to modify the primer scheme bed files so that all positions affected by this mispriming are trimmed. As amplicons 63 and 64 have a large overlap this will not introduce a gap to the end consensus sequence, as can be seen in the primer-trimmed BAM (Figure 4). This change has been tested using the ARTIC fieldbioinformatics and ncov2019-artic-nf pipelines.
Figure 4