Epidemiological and phenotypic characterization of certain variants of SARS-CoV-2 have highlighted the changing transmissibility, infectivity and vaccine escape capability of this virus. Of considerable interest are the B.1.351 variant (20H/501Y.V2), and B.1.1.7 variant (20I/501Y.V1) that have now been reported from multiple countries around the world. To track the evolution of SARS-CoV-2 in Bangladesh, the Child Health Research Foundation (CHRF) sequenced 161 SARS-CoV-2 genomes and identified the circulating variants. We have also developed a country-specific build in Nextstrain of all SARS-CoV-2 isolates sequenced and uploaded on GISAID from Bangladesh (auspice). On 23 March 2021, we uploaded a tree containing 835 Bangladeshi SARS-CoV-2 genome sequences contextualized against 3,653 genomes across the globe.
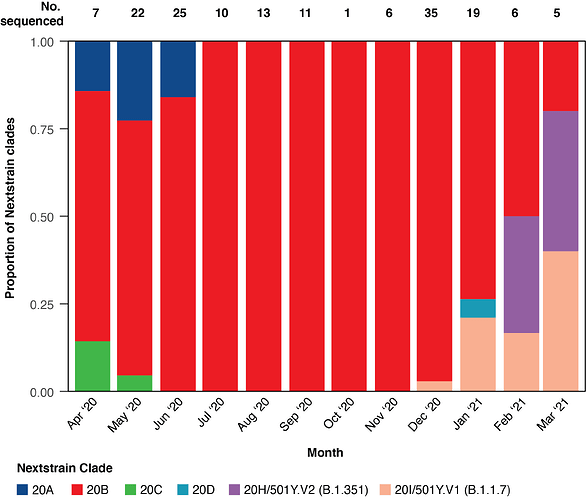
Figure 1: Monthly distribution of Nextstrain clades of 161 SARS-CoV-2 genomes sequenced at the Child Health Research Foundation (CHRF) in Bangladesh from April 2020 to March 2021. The number of genomes sequenced per month are also included.
The monthly distribution of the Nextstrain clades of SARS-CoV-2 sequences from our testing facility are shown in Figure 1. The first B.1.1.7 variant was identified in December 2020 and since then, 7 additional sequences from this clade were detected. The first sequenced B.1.351 variant from Bangladesh was detected in a patient on January 24, 2021. We have captured four B.1.351 variants in our surveillance starting from February 27, 2021. In March 2021, 4/5 of SARS-CoV-2 genomes sequenced belonged to either B.1.1.7 or B.1.351.
Conclusions
This report highlights the increasing isolation of the B.1.1.7 and B.1.351 variants in Bangladesh. The B.1.1.7 variant was first detected in December 2020 and since then ~23% of all genomes sequenced belong to this variant. The B.1.351 variant was first identified in Bangladesh in January 2021, and here we report four additional genomes belonging to this variant in the last two months. The isolation of these variants correlates with an increase in the number of SARS-CoV-2 cases and test positivity rate in Bangladesh. Genomic surveillance must continue to monitor this trend as these findings have grave implications for mitigation and vaccination policies. These findings are of utmost importance to policy makers who need to relay this information to the public and start instituting appropriate public health policies. If this trend continues, we recommend re-introduction of non-pharmaceutical interventions to prevent further spread of these variants within Bangladesh and rise of new ones. In addition, vaccination efforts much be continued to limit infection rates.
The findings should be considered within the context of a few limitations. First, majority of the samples were received at CHRF from specific hospitals/testing centers, and therefore the results may be biased by local outbreaks. Second, the sequenced genomes are a small proportion of confirmed SARS-CoV-2 cases, and hence may not reflect the diversity of SARS-CoV-2 circulating in Bangladesh. Despite these limitations, detection of these variants is very concerning and warrants immediate attention of policy makers and public health officials.
Data availability
All the SARS-CoV-2 sequences from CHRF are available on GISAID.
Partners and Collaborators
Child Health Research Foundation, Dhaka, Bangladesh
Disclaimer and contact information
Please note that these analyses are based on ongoing work and should be considered preliminary. For further information, please contact:
Senjuti Saha, PhD
Scientist, Child Health Research Foundation, Dhaka, Bangladesh
E-mail: [email protected]
Arif Mohammad Tanmoy, MS
Research Investigator, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Syed Muktadir Al Sium, MS
Molecular Biologist, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Afroza Akter Tanni, MS
Molecular Biologist, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Sharmistha Goswami, MS
Microbiologist, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Hafizur Rahman, DMT
Senior Research Technologist, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Samir Saha, PhD
Executive Director, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Yogesh Hooda, PhD
Visiting Scientist, Child Health Research Foundation, Dhaka, Bangladesh
Email: [email protected]
Acknowledgements
We thank the Director General Health Services, Government of Bangladesh, and the Institute of Epidemiology, Disease Control, and Research for giving us the opportunity to work as a COVID-19 response team. We thank the health facilities in Bangladesh that sent us samples for COVID-19 testing and all personnel at CHRF for their work at the COVID-19 diagnostic laboratory. We are grateful to Shimantik Pathology and Diagnostic Center, Syhlet for providing us with samples for sequencing. We would like to thank the Chan Zuckerberg Biohub, the Chan Zuckerberg Initiative and the Quadram Institute for their intellectual support and guidance.
We received funding from the Bill and Melinda Gates Foundation (grant INV-016932) for this project.