Background
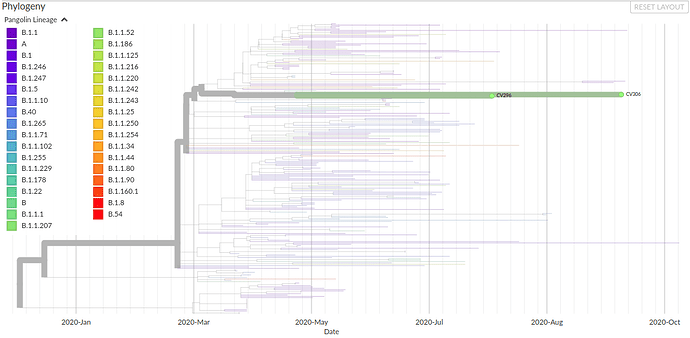
Rambaut et al. published a report on the 19th of December 2020 describing a preliminary genomic characterization of an emergent SARS-CoV-2 lineage (B.1.1.7) in the United Kingdom and emphasized the need for an enhanced genomic and epidemiological surveillance of the lineage worldwide. Following the release of the report, we did a quick analysis of samples recently sequenced at the African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Nigeria and we identified two SARS-CoV-2 sequences that share one non-synonymous SNP in the spike protein in common with this lineage, but none of the 22 other unique lineage-defining mutations of B.1.1.7. These were found in two patient samples collected on the 3rd of August 2020 and 9th of October 2020 from individuals in Osun State, Nigeria. The non-synonymous SNP, S:P681H, has been observed in global data outside of the B.1.1.7 lineage, and likely represents an independent, homoplasic occurrence of this substitution from the UK.
Figure 1: Phylogeny of SARS-CoV-2 genomes from Nigeria showing a distinct clade of the B.1.1.207 lineage with branch length indicating sample collection dates. The two genomes were detected in Osun state, Nigeria, with collection dates on 3rd of August and 9th of October, 2020.
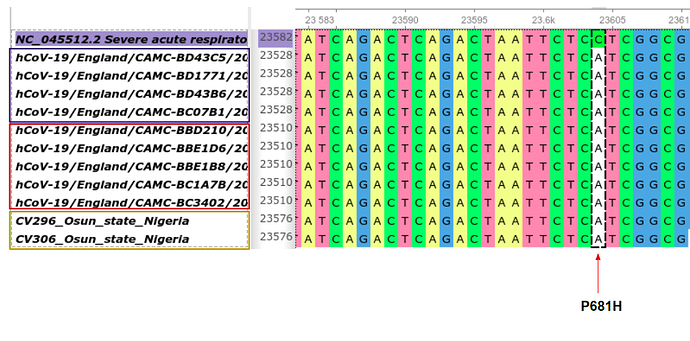
Figure 2: Multiple alignment of SARS-CoV-2 reference genome with genomes from Osun state, Nigeria and the United Kingdom. Only two genomes (highlighted in yellow) in Nigeria (Osun state) have been seen to have the P681H mutation so far. This non-synonymous mutation is found in the B.1.1.7 lineage (highlighted in red) and can also be found in other UK lineages such as the B.1.255 (highlighted in purple).
The P681H spike protein mutation has been previously noted to be directly adjacent to the furin cleavage site, which may have biological significance. However, unlike the B.1.1.7 lineage described in the UK, we haven’t observed such rapid rise of the lineage in Nigeria and do not have evidence to indicate that the P681H variant is contributing to increased transmission of the virus in Nigeria. However, the relative difference in scale of genomic surveillance in Nigeria vs the UK may imply a reduced power to detect such changes; we are currently in the process of sequencing more samples from the recent surge in the number of cases of covid-19 in Nigeria.
Summary
At the moment, only about 1% of the SARS-CoV-2 genomes from Nigeria share any of the 17 protein-altering variants from the UK lineage of concern (B.1.1.7). However, this might change in the next few weeks when we sequence more samples from the recently reported surge of covid-19 in Nigeria. Other reported mutations such as the N501Y, A570D, and the HV 69 - 70 deletion in the spike protein have not been detected in our genomes yet, this could also be associated with non-increase of the lineage in Nigeria currently. Inasmuch as the full effect of this mutation isn’t known yet and it is still being studied, the importance of a robust genomic surveillance system cannot be overemphasized as we can identify and report changes in the genomes of pathogens such as SARS-CoV-2 that are of public health interest.
Data availability
All our Nigerian SARS_CoV_2 sequences obtained from the beginning of the pandemic in Nigeria till date are available at CoV_Sequences/all_nigeria_sars-cov-2_sequences_12-21-20.fasta at master · acegid/CoV_Sequences · GitHub. All of these sequences are also available on GISAID.
Partners and Collaborators
African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Ede, Osun State, @acegid.
Redeemer’s University, Ede, Osun State (RUN), @RedeemersUni
Nigeria Centre for Disease Control (NCDC), Abuja, Nigeria, @NCDCgov.
Africa Center for Diseases Control and Prevention (AfCDC), @AfricaCDC.
Disclaimer and contact information
Please note that these analyses are based on work in progress and should be considered preliminary. Our analyses of this data are ongoing and a publication communicating our findings on these and other published genomes is in preparation. These data cannot be used without permission. If you wish to use this data please contact:
Christian Happi, PhD
Professor of Molecular Biology and Genomics, Redeemer’s University, Ede, Osun State, Nigeria
Director, African Center of Excellence for Genomics of Infectious Diseases [ACEGID]
E-mail: [email protected]
Website: www.acegid.org
Twitter: @christian_happi
Chikwe Ihekweazu, M.P.H, F.F.P.H
Director-General, Nigerian Centre for Disease Control [NCDC], Abuja, Nigeria
Email: [email protected]
Website: https://ncdc.gov.ng
Twitter: @Chikwe_I
John Nkengasong, PhD
Director-General, Africa Centre for Disease Control and Prevention (AfDCD)
Addis-Abeba, Ethiopia
Email: [email protected]
Twitter: @JNkengasong
Paul Eniola Oluniyi, MSc, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
E-mail: [email protected]
Twitter: @pauloluniyi
Idowu Olawoye, MSc, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
Email: [email protected]
Twitter: @idowuolawoye