I have done a root-to-tip regression using the latest build from nextstrain (ML by iqtree, rooted in treedater). The results are not very compelling:
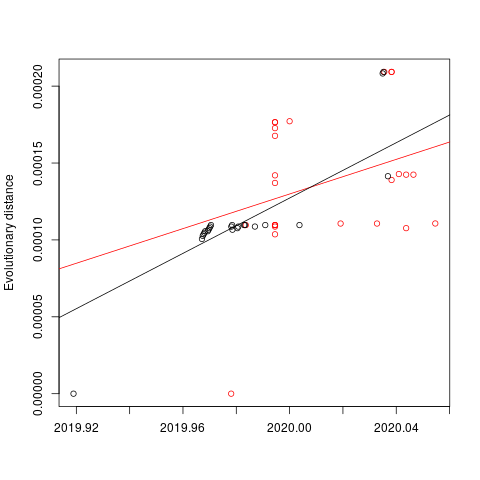
Root-to-tip mean rate: 0.00056301772302758
Root-to-tip p value: 0.139650296902652
Root-to-tip R squared (variance explained): 0.0923786229950628
treedater is not able to get a stable estimate of the rate with these data. If I fix the rate at 8e-4 I estimate the following dates:
"2019-12-02 09:54:53 UTC"
2.5% 97.5%
"2019-11-02 22:07:58 UTC" "2019-12-16 09:36:07 UTC"
I suspect that getting robust & precise estimates of the TMRCA will require more samples, ideally including the animal reservoir, but having a better estimate of the rate would help a lot too.