Following up on our sequencing efforts during the Nigeria 2018 Lassa fever outbreak, we here present the first sequences from our response to the Nigeria 2019 Lassa fever outbreak, in a collaboration between the Irrua Specialist Teaching Hospital (ISTH), Institute for Lassa Fever Research and Control (ILFRC), Irrua, Edo State, Nigeria; the Bernhard-Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany; the Rega Institute / KU Leuven, Leuven, Belgium; the ARTIC network; and Public Health England (PHE), Salisbury, United Kingdom.
In order to obtain the sequences for these 19 samples, MinION technology (Oxford Nanopore) is being used in conjunction with a non-targeted metagenomic RNA sequencing approach, which facilitates detection and sequencing of known pathogens including new lineages or clades of known pathogens as well as unknown pathogens. We refer to Kafetzopoulou et al. for more information (1).
We here make available sequence data from 19 laboratory-confirmed Lassa fever patients, with sampling dates ranging from December 31st 2018 until February 2nd 2019 (additional sequences are in the pipeline). Please note this is an early release with preliminary data (see also disclaimer below). These 19 Lassa virus sequences were generated within two weeks of set-up and implementation of the sequencing laboratory at ISTH, and originate from the States of Edo (9), Ondo (9) and Kogi (1) and their consensus sequences for both the L and S segments can be downloaded here:
https://github.com/ISTH-BNITM-KUL-PHE/LASVsequencing2019
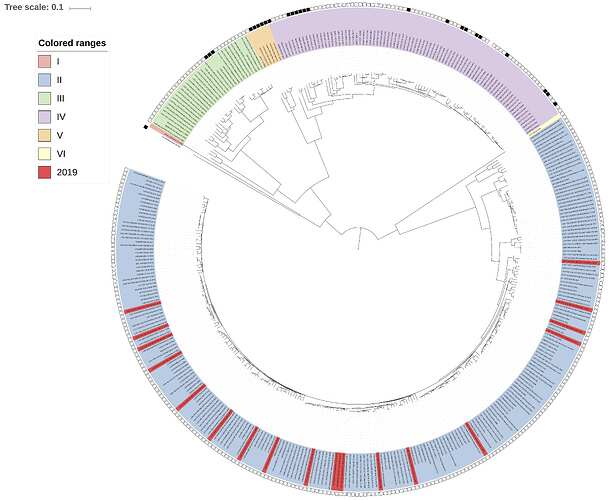
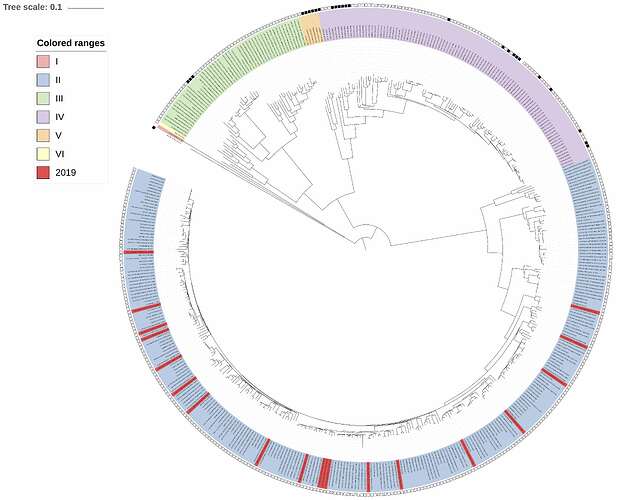
Complementing the 19 newly-obtained sequences, we performed maximum-likelihood phylogenetic analysis of the coding genes in both the L and S segments, across all LASV lineages (I-VI), including those of 2018 and earlier years published by Kafetzopoulou et al. in Science in early January 2019 (1). Each of the new sequences was placed with the genotype II lineage, in both the L and S segments analyses (Figures 1 and 2, respectively). Thus, these 19 viruses transmitted in 2019 originate from the pool of lineages, sub-lineages and strains known to circulate in Nigeria. These results are in line with observations made during the 2018 Lassa fever outbreak.
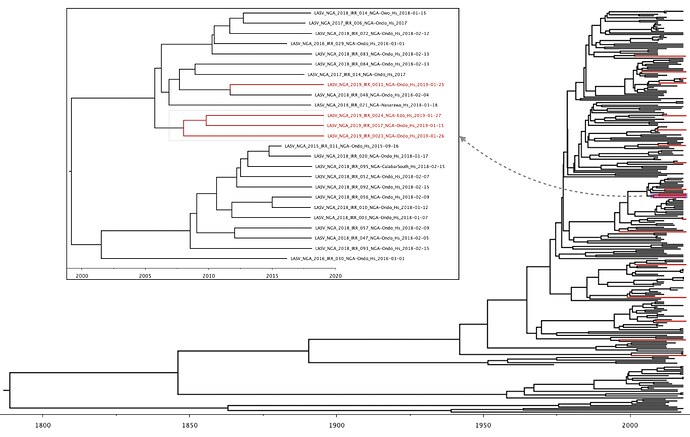
In both L and S segment analyses, a single cluster of three 2019 viruses was observed (LASV-NGA-2019-IRR-0017, LASV-NGA-2019-IRR-0023 & LASV-NGA-2019-IRR-0024). In order to estimate the time of most recent common ancestors (tMRCAs) across the tree, we are currently performing BEAST analysis on the genotype II lineage of the S segment data. These analyses are still ongoing but indicate a preliminary tMRCA for this cluster dating back to the second half of 2007 (95% highest posterior density: 2005-2010; see Figure 3). Other divergence time estimates are similar to those of Kafetzopoulou et al. (1). This preliminary analysis hence shows infection events with different viruses that are not directly linked. This is consistent with spillover of viruses from the rodent reservoir to humans rather than extensive human-to-human transmission.
Figure 1: Lassa virus L-segment maximum-likelihood phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different colour ranges. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. Newly sequenced viruses are depicted in red. All newly sequenced samples reside within the genotype II clade.
Figure 2: Lassa virus S-segment maximum-likelihood phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different colour ranges. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. Newly sequenced viruses are depicted in red. As with the L-segment phylogeny, all newly sequenced samples reside within the genotype II clade.
Figure 3: Lassa virus S-segment time-measured phylogeny (preliminary result), focusing on the genotype II lineage. Newly sequenced viruses are depicted in red. A single clade containing three 2019 Lassa virus sequences is shown in more detail.
Platform for sharing of sequences
The unpublished sequences are shared via the website virological.org under the following conditions: The Irrua Specialist Teaching Hospital (ISTH), Edo State, Nigeria, Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany, and Public Health England (PHE), UK, are committed to sharing data in public health emergencies. We release unpublished Lassa virus sequences from Nigeria to support the public health response as well as the development and evaluation of Lassa fever diagnostics or therapeutics. The data may be used and analyzed for these purposes. It is not permitted to use the sequences for publication, i.e. any type of communication with the general public that describes data generated with the use of the sequences. If you intend to so, please contact us directly:
Dr. Ephraim Ogbaini <[email protected]>, Director of Institute of Lassa Fever Research and Control,
Prof. Stephan Günther <[email protected]>, Director, WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Reference and Research, BNITM
(1) L. E. Kafetzopoulou, S. T. Pullan, P. Lemey, M. A. Suchard, D. U. Ehichioya, M. Pahlmann, A. Thielebein, J. Hinzmann, L. Oestereich, D. M. Wozniak, K. Efthymiadis, D. Schachten, F. Koenig, J. Matjeschk, S. Lorenzen, S. Lumley, Y. Ighodalo, D. I. Adomeh, T. Olokor, E. Omomoh, R. Omiunu, J. Agbukor, B. Ebo, J. Aiyepada, P. Ebhodaghe, B. Osiemi, S. Ehikhametalor, P. Akhilomen, M. Airende, R. Esumeh, E. Muoebonam, R. Giwa, A. Ekanem, G. Igenegbale, G. Odigie, G. Okonofua, R. Enigbe, J. Oyakhilome, E. O. Yerumoh, I. Odia, C. Aire, M. Okonofua, R. Atafo, E. Tobin, D. Asogun, N. Akpede, P. O. Okokhere, M. O. Rafiu, K. O. Iraoyah, C. O. Iruolagbe, P. Akhideno, C. Erameh, G. Akpede, E. Isibor, D. Naidoo, R. Hewson, J. A. Hiscox, R. Vipond, M. W. Carroll, C. Ihekweazu, P. Formenty, S. Okogbenin, E. Ogbaini-Emovon, S. Günther, S. Duraffour; Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak; Science 04 Jan 2019: 74-77; DOI: 10.1126/science.aau9343