Following up on our previous release of in total 42 sequences from our response to the Nigeria 2019 Lassa fever outbreak (see our previous posts on virological.org), we here present an additional set of 33 samples Lassa virus draft sequences, bringing the total number of samples sequenced to 75. They are derived from 75 laboratory-confirmed Lassa fever patients from Edo, Ondo, Kogi, Delta, Ebonyi and Benue States with dates of onset ranging from the end of December 2018 to the beginning of March 2019. These sequences were generated through a collaboration between the Irrua Specialist Teaching Hospital (ISTH), Institute for Lassa Fever Research and Control (ILFRC), Irrua, Edo State, Nigeria; the Bernhard-Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany; the Rega Institute / KU Leuven, Leuven, Belgium; the ARTIC network; and Public Health England (PHE), Salisbury, United Kingdom.
Of the 75 sequences, 73 sequences fall within lineage II viruses and one (2019_IRR_0070) falls within lineage III viruses which are known to circulate in these states. One “divergent strain” (2019_IRR_082), which may potentially represent a new lineage, has been detected in Ondo State, at the border of Ekiti State. This outlier strain seems to cluster with one virus previously sequenced from a human plasma sample from Ekiti State (i.e. as part of another project, D. Ehichioya et al. , manuscript in preparation) and also appears quite distantly related to the Kak-428-2012 strain which was sequenced from one of the new hosts of Lassa virus, the African wood mouse Hylomyscus pamfi in Nigeria (1). While the detection of these “divergent strains” may be seen as a rare event as only two strains have been detected so far (i.e., in 2016 and 2019), future surveillance in the affected states will enable to gain more knowledge about the emergence of this potential new lineage. Further bioinformatics analyses are ongoing to investigate this finding.
In order to obtain these sequences, MinION technology (Oxford Nanopore) is being used in conjunction with a non-targeted metagenomic RNA sequencing approach, which facilitates detection and sequencing of known pathogens including new lineages or clades of known pathogens as well as unknown pathogens. We refer to Kafetzopoulou et al. for more information (2). These 75 Lassa virus sequences were generated within four weeks of set-up and implementation of the sequencing laboratory at ISTH.
We here make available sequence data from an additional 33 laboratory-confirmed Lassa fever patients, with sampling dates ranging from December 31st 2018 until March 3rd 2019 (additional sequences are in the pipeline). Please note this is an early release with preliminary data (see also disclaimer below). Their consensus sequences for both the L and S segments can be downloaded here:
https://github.com/ISTH-BNITM-KUL-PHE/LASVsequencing2019
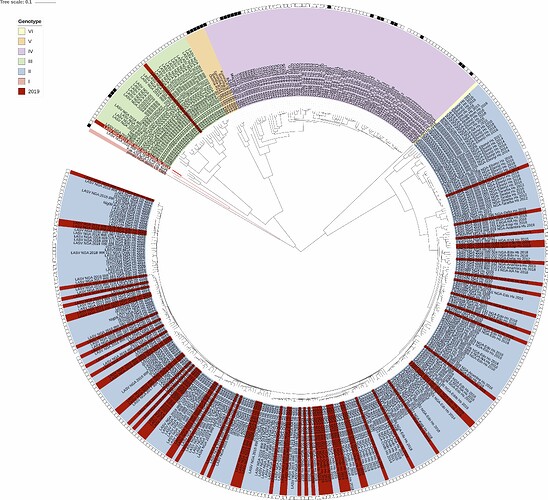
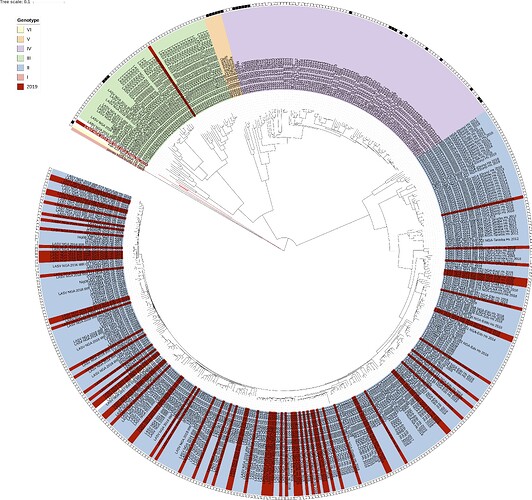
We again performed maximum-likelihood phylogenetic analysis of the coding genes in both the L and S segments, across all LASV lineages (I-VI), including those of 2018 and earlier years published by Kafetzopoulou et al. in Science in early January 2019 (1). The 75 sequences represent a total of 63 complete S and L sequences, and 12 partials with S-segment only. As shown in Figures 1 and 2, maximum-likelihood phylogenetic analyses of the coding genes in both S and L segments indicates spill-over from the rodent hosts within lineages II and III (74 sequences). The “divergent strain” (2019_IRR_082), which may potentially represent a new lineage, originates from a sample from Ondo State, at the border of Ekiti State.
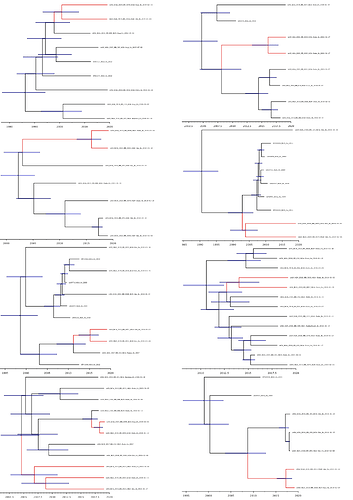
In order to estimate the time of most recent common ancestors (tMRCAs) across the tree, we have generated a time-measured maximum clade credibility tree using BEAST on the genotype II lineage of the S segment data (Figure 3). The tMRCAs of the eight clusters of newly sequenced viruses in 2019 show infection events with different viruses that are not directly linked. This is consistent with spill-over of viruses from the rodent reservoir to humans rather than extensive human-to-human transmission. Also, the detection of this “divergent strain” may be seen as a rare event and future surveillance in the affected States will enable to gain more knowledge about this. Thus, the vast majority of the viruses transmitted in 2019 originate from the pool of lineages, sub-lineages and strains known to circulate in Nigeria.
Additional sequences may be shared following sequencing at ISTH and analysis following return back to Europe. The sequencing team is still working closely with clinicians and epidemiologists to analyse potential human clusters. However, this remains challenging as samples with low viremia do not allow for recovery of full genome sequences (S and L) which thus renders comparison between potential human cluster samples difficult. Efforts are being made into this and results will be updated accordingly.
Figure 1: Lassa virus L-segment maximum-likelihood phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different colour ranges. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. Newly sequenced segments generated in 2019 (63 in total so far) are depicted in red. Apart from 2 sequences (see above), all newly sequenced samples reside within the genotype II clade.
Figure 2: Lassa virus S-segment maximum-likelihood phylogeny. The circular tree includes all LASV lineages (I-VI) represented by different colour ranges. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. Newly sequenced segments generated in 2019 (75 in total so far) are depicted in red. As with the L-segment phylogeny, apart from 2 sequences (see above), all newly sequenced samples reside within the genotype II clade.
Figure 3: Lassa virus S-segment time-measured phylogeny, focusing on the genotype II lineage. Eight clusters of newly sequenced viruses are depicted in red.
Platform for sharing of sequences
The unpublished sequences are shared via the website virological.org (github link) under the following conditions: The Irrua Specialist Teaching Hospital (ISTH), Edo State, Nigeria; Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany; Rega Institute, KU Leuven, Leuven, Belgium; the ARTIC network; and Public Health England (PHE), UK, are committed to sharing data in public health emergencies. We release unpublished Lassa virus sequences from Nigeria to support the public health response as well as the development and evaluation of Lassa fever diagnostics or therapeutics. The data may be used and analyzed for these purposes. It is however not permitted to use the sequences for publication. If you intend to so prior to our publication, please contact us directly:
Dr. Ephraim Ogbaini <[email protected]>, Director of Institute of Lassa Fever Research and Control,
Prof. Stephan Günther <[email protected]>, Director, WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever Reference and Research, BNITM
(1) Olayemi A, Cadar D, Magassouba N, Obadare A, Kourouma F, Oyeyiola A, Fasogbon S, Igbokwe J, Rieger T, Bockholt S, Jérôme H, Schmidt-Chanasit J, Garigliany M, Lorenzen S, Igbahenah F, Fichet JN, Ortsega D, Omilabu S, Günther S, Fichet- Calvet E; New Hosts of The Lassa Virus. Sci Rep. 2016; 6:25280. doi: 10.1038/srep25280.
(2) L. E. Kafetzopoulou, S. T. Pullan, P. Lemey, M. A. Suchard, D. U. Ehichioya, M. Pahlmann, A. Thielebein, J. Hinzmann, L. Oestereich, D. M. Wozniak, K. Efthymiadis, D. Schachten, F. Koenig, J. Matjeschk, S. Lorenzen, S. Lumley, Y. Ighodalo, D. I. Adomeh, T. Olokor, E. Omomoh, R. Omiunu, J. Agbukor, B. Ebo, J. Aiyepada, P. Ebhodaghe, B. Osiemi, S. Ehikhametalor, P. Akhilomen, M. Airende, R. Esumeh, E. Muoebonam, R. Giwa, A. Ekanem, G. Igenegbale, G. Odigie, G. Okonofua, R. Enigbe, J. Oyakhilome, E. O. Yerumoh, I. Odia, C. Aire, M. Okonofua, R. Atafo, E. Tobin, D. Asogun, N. Akpede, P. O. Okokhere, M. O. Rafiu, K. O. Iraoyah, C. O. Iruolagbe, P. Akhideno, C. Erameh, G. Akpede, E. Isibor, D. Naidoo, R. Hewson, J. A. Hiscox, R. Vipond, M. W. Carroll, C. Ihekweazu, P. Formenty, S. Okogbenin, E. Ogbaini-Emovon, S. Günther, S. Duraffour; Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak; Science 04 Jan 2019: 74-77; DOI: 10.1126/science.aau9343