Report by Nuno Faria on behalf of the ZiBRA sequencing consortium
Cross-post from: http://zibraproject.github.io/blog/preliminary-results-from-lab-work/
Data collected during the ZiBRA road trip
After a short but intense journey through Zika virus hotspots, the first phase of the ZiBRA project came to an end on the 17 June in Salvador, Bahia. After the “road trip” (phase 1 of the project) the ZiBRA team has been busy analysing data, optimizing protocols and applying for funding to continue and expand this venture in Brazil. In the meanwhile, it is worth having a look at the data collected during the 2-week trip.
Very early on during our road trip we realized that no ZIKV RNA had been extracted in the LACENs (Laboratório Central de Saúde Pública), and confirmation by an RT-PCR assay needed to be done for all samples analysed throughout the trip using a standardized protocol (we followed the Lanciotti protocol). This would guarantee that all samples had been tested using the same assay and, perhaps most importantly, it also meant that we could deliver the results of yet untested samples back to the LACENs. LACENs typically send their samples to a national lab, but unfortunately results either arrive 9 months later or simply don’t arrive. With the tireless efforts of our teams from University of Sao Paulo, FioCruz Bahia and Evandro Chagas and with the valuable help of local laboratories, we managed to test an average of 300 samples over 2-day periods and the results were delivered within 48h upon our arrival to each laboratory. Also, because we obtained positive results for several samples that had tested negative in previous tests done by some LACENs, we were able to propose a more accurate diagnostic of Zika virus for future surveillance. Formal reports of the ZiBRA activities in each LACEN were prepared by Luiz Alcantara and sent to the Ministry of Health (MoH).
Table 1 summarizes some characteristics of the samples obtained during the road trip. In representative locations (LACENs), we found between 8-12% positive samples, with mean Ct values of 36 (min: 18.6 to max: 42.1). We also found that the lag between onset of symptoms and date of collection tends to be shorter for PCR-positive samples, with a median of 2 (0, 34) days and 4 (0, 115) days respectively for positive and negative samples. Similar numbers have been reported for the Zika epidemic in Colombia. This scenario is also in line with recent studies done on rhesus macaque models showing that Zika viral loads tend to be highest 2-3 days after infection and decline rapidly after the onset of symptoms.
[Table 1] Characteristics of the human samples analysed during the ZiBRA road-trip. Green arrows indicate sampling done at the LACENs, while grey arrows denote convenience sampling.
It is worth here to praise the tremendous effort from LACEN teams in coping with the epidemics of dengue, and then the rampant epidemics of Chikungunya and Zika virus. In 2015, most cases potentially presenting with Zika virus infection where not attributed a GAL identifier number and were not included in the SINAN system. This made the retrospective identification of date of sample collection and municipality of residence somewhat more challenging - but not impossible in most cases. For example, we found the 2 earliest positive cases from Brazil that have been described so far. The first reports of Zika autochthonous cases in Brazil were from patients residing in Camaçari (a middle-sized city with a strong petrochemical industry located < 1h from the capital of Bahia, Salvador; published here http://wwwnc.cdc.gov/eid/article/21/10/15-0847_article) and from Natal (capital of Rio Grande do Norte; report in http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4501423/). Both reports indicate that earliest infections were detected in March 2015. During our visit to the first LACEN in Natal, we found two RT-PCR positive samples collected in February 2015. These “historical” samples are, so far, the earliest samples identified in Brazil and should contribute to a better understanding of the ignition and early spread of this virus in the country.
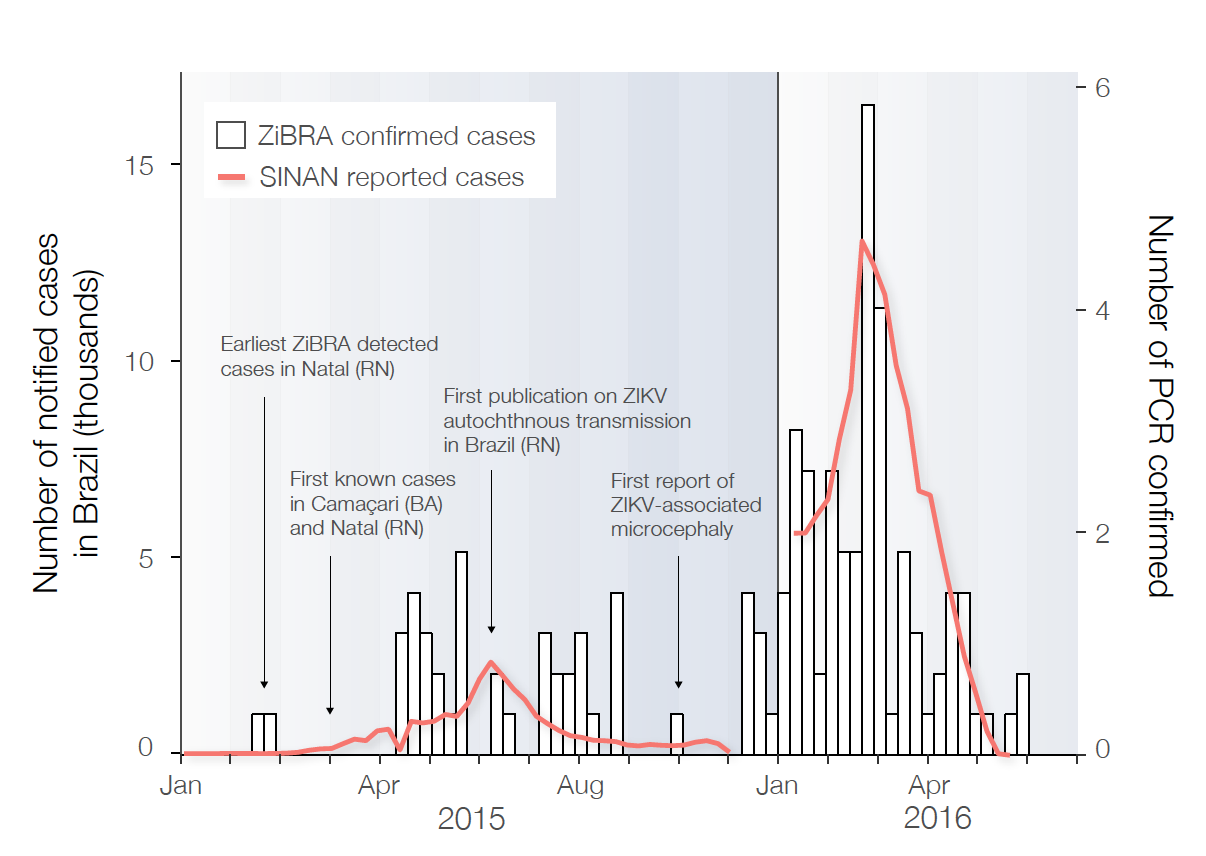
More generally, the MoH contributed significantly to our efforts and provided us with Zika notified cases per week for each municipality and federal state in 2016. This allowed us to focus on testing samples that had been collected during the reported epidemic peaks at each state. Figure 2 shows the date of collection of the positive samples collected during the road trip (bars) with the reported SINAN cases from weeks 1-49 2015 (reported in http://science.sciencemag.org/content/early/2016/03/23/science.aaf5036) and the new SINAN data from 2016 (in red). Notably both the RT-PCR confirmation and the SINAN notified cases indicate two waves of Zika virus in Brazil, one peaking in July 2015, and another wave, possibly even greater in absolute numbers of infected individuals, in the end of February 2016. Also, it is interesting to note that federal states with higher incidence of ZIKV (and microcephaly) incidence tend to have higher percentage of positive samples.
[Figure 1] Number of PCR-confirmed cases of ZIKV in human samples collected by ZiBRA (white bars) and number of notified cases by the Ministry of Health (red line).
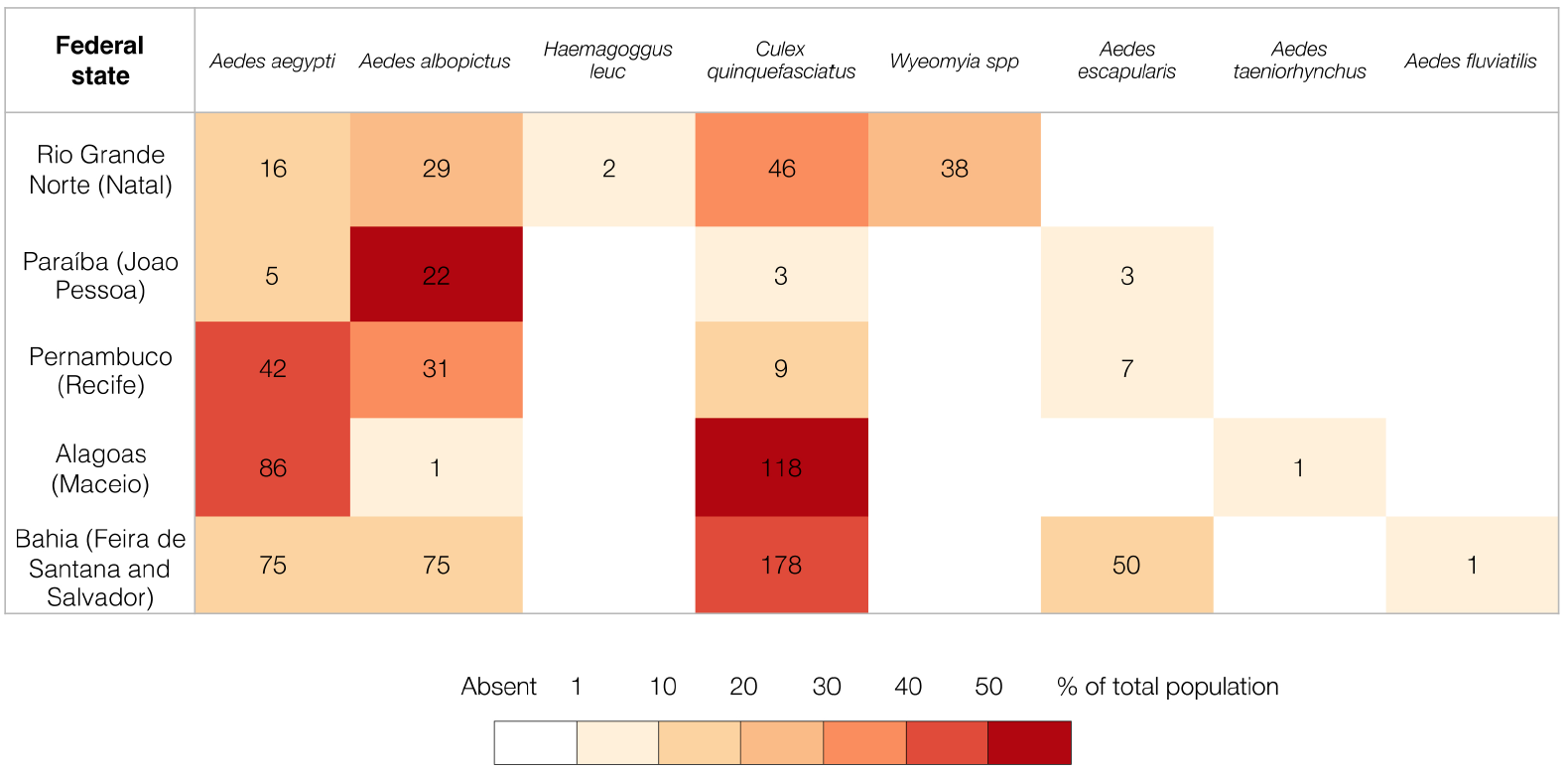
Finally, the entomology team lead by Poliana Lemos, Bruna Nascimento and Hamilton Monteiro, and helped by volunteers from local Epidemiological Surveillance teams, captured a total of 838 mosquitoes in 19 collection points across 5 federal states. A total of 8 different species were identified. Aedes aegypti, Aedes albopictus and Culex quinquefasciatus were detected in all locations. Other mosquito species were also detected. For example, we detected the Haemagogus leucocelaenus in Natal, a vector typically involved in enzootic outbreaks of yellow fever, and also the less common Aedes fluviatis and taeriorhynchus. RNA extracted from these has now been pooled at the Technology Innovation Center, Instituto Evandro Chagas and virus detection and mosquito sequencing are currently being performed.
[Figure 2] Mosquitoes captured during the ZiBRA road trip. Colour gradient indicates the proportion of a particular mosquito species at a given location.
To conclude, positive samples as well as sequencing reagents have been split evenly between the participating labs in Brazil (Ester Sabino’s team at the University of Sao Paulo, Luiz Alcantara’s team at FioCruz Bahia and Marcio Nunes team at the CIT, Evandro Chagas Institute). We also would like to welcome our new ZiBRA partners at FioCruz Recife (team’s of Rafael Dhalia http://www.cpqam.fiocruz.br/?option=com_pplp&Itemid=5&action=pesquisadorShowDetalhe&id=15 and Constancia Ayres http://www.cpqam.fiocruz.br/?option=com_pplp&Itemid=5&action=pesquisadorShowDetalhe&id=17). Soon to follow more on the sequencing low Ct value samples and phylogenetic analysis of sequenced isolates.