Update information including data from week 3, 2020
Weekly proportion of SGTF and SGTL positive samples among all TaqPath positive samples detected, between week 49 (2020) and week 3 (2021), by a large laboratory distributed throughout the Portuguese territory (n = 36651 TaqPath positive samples).
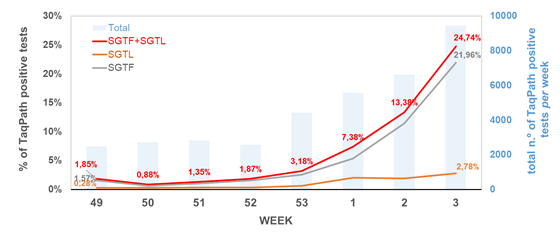
Figure notes
SGTF (S gene target failure) = positive test with non-detectable S gene and <=30 Cycle threshold (Ct) for N and ORF1ab targets; SGTL (S gene target late detection) = positive test having Ct values for S gene >5 units higher than the maximum Ct value obtained for the other two targets (N and ORF1ab) of the assay (exclusively includes positive samples with <=30 Ct for N and ORF1ab targets). The mean Ct difference between S gene and N/ORF genes for SGTL samples was consistently around 5-8 Ct values (median=6.60, IQR=5.79-7.59)
Forecast for weeks 4 to 6 (2021) of the estimated proportion of SGTF/SGTL and B.1.1.7 cases
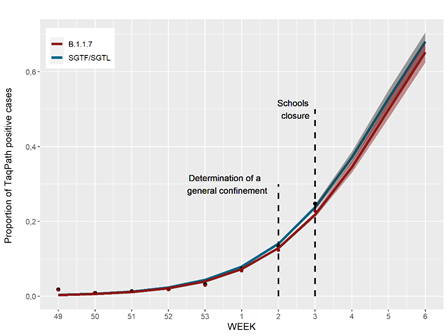
Figure notes
Given that the increase SGTF/SGTF and B.1.1.7 frequency likely tends to plateau (i.e., the increasing rate tends to reduce with time), we opted to model the weekly trend of the SGTF/SGTF and B.1.1.7 frequency with a logistic model (instead of log-binomial model used at week 2). The proportion of SGTF/SGTL cases that are B.1.1.7 was estimated as 0.918 (0.845 to 0.964, CI 95%) based on a sub-sample of sequencing cases (90 B.1.1.7 cases in 98 known SGTF/SGTL sequenced samples analysed so far).