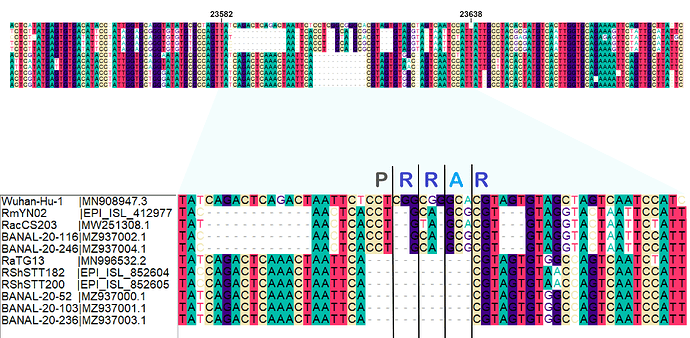
We have previously proposed that the furin cleavage site (FCS) in the SARS-CoV-2 Spike protein - a feature absent from the closest known relatives of SARS-CoV-2 for that genomic region - was inserted through a recent polymerase copy-choice error between the progenitor of SARS-CoV-2 and a co-circulating Sarbecovirus (post above). We observed evidence for sequence homology between the SARS-CoV-2 FCS nucleotide sequence and the respective genomic region in RmYN02, a virus closely related to SARS-CoV-2 for most of its genome apart from the 5’ end of the Spike gene, including the FCS.
Following significant virus sampling efforts and re-analysis of previous samples since the start of the pandemic, we now know of 3 more viruses that share the RmYN02-like sequence for this region of Spike: RacCS203 sampled in Thailand in a R. acuminatus bat (Wacharapluesadee et al., 2021), BANAL-20-116 and BANAL-20-247 both sampled in Laos in R. malayanus bats (Temmam et al., 2021). There are also an additional five viruses with the SARS-CoV-2-like sequence, but missing the FCS: RShSTT182 and RShSTT200 sampled in Cambodia in R. shameli bats (Delaune et al., 2021), and BANAL-20-52, BANAL-20-103, BANAL-20-236 sampled in Laos in R. malayanus, pusillus and marshelli bats respectively (Temmam et al., 2021).
Figure 1. Nucleotide sequence alignment of SARS-CoV-2 bat CoV relatives at the genomic region of the FCS (Wuhan-Hu-1 coordinates: 23582-23638).
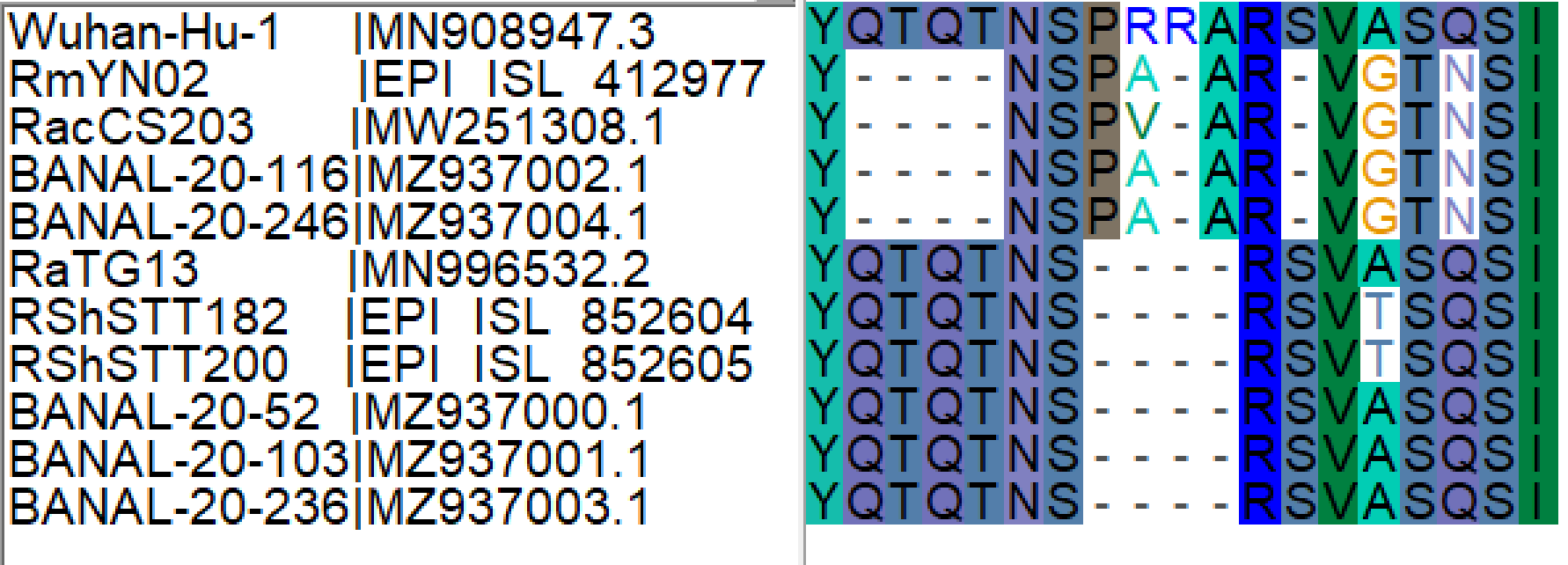
The clear homology of these newly discovered viruses (Figures 1 and 2) reinforces our previous observation that the RmYN02-like lineage (referred to as Clade X in the original post) for this genomic region is the likely origin of the SARS-CoV-2 FCS through copy-choice error in a mixed infection.
Figure 2. Protein sequence alignment of SARS-CoV-2 bat CoV relatives at the genomic region of the FCS (Wuhan-Hu-1 Spike protein coordinates: 674-692).
This scenario would of course require co-circulation of viruses with both sequences in the same host population. The discovery of the five BANAL-20 viruses (two with the RmYN02-like sequence and five with the SARS-CoV-2-like sequence), all sampled in the same site (Fueng district in Vientiane Province, Laos), provides clear evidence of both genomic backgrounds infecting the same bat populations and increases the likelihood of our original hypothesis for the SARS-CoV-2 FCS origin.
A final - arguably rather speculative - clue supporting this hypothesis comes from 2 nucleotides in the alignment. The sequence of RacCS203 matching the SARS-CoV-2 FCS is more similar to that of SARS-CoV-2 than the other 3 RmYN02-like sequence (RmYN02, BANAL-20-116, BANAL-20-247) by a single A (gcAcgt) instead of a G that is present in the latter (gcGcgt). The presence of this A in the RacCS203 sequence coincides with the presence of a T on the third position shown in this alignment. This T is shared between RacCS203, SARS-CoV-2 and the other 4 viruses closest to SARS-CoV-2 in that region - RaTG13, BANAL-20-52, BANAL-20-103 and BANAL-20-236 - while a C is present in that position in the viruses that have a G instead of an A in the aforementioned position. The consistent homology for these two SNPs could be evidence that the RmYN02-like sequence is indeed phylogenetically related to the SARS-CoV-2 FCS sequence and not just due to convergent sequence homology. Nevertheless, this SNP homology is in no way conclusive, since both changes are transitions (G - A, T - C) that could have taken place convergently between RacCS203 and SARS-CoV-2 (/the clade X virus that recombined with SARS-CoV-2 to insert the pre-FCS sequence).
Figure 3. Co-occurring SNPs in the potentially phylogenetically homologous region to the SARS-CoV-2 FCS.
It is still worth noting once more that the more of these viruses we sample, the clearer it will be how ‘unique’ (or how common) the SARS-CoV-2 FCS actually is in nature.
Spyros Lytras
References
Delaune, D. et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 2021 121 12, 1–7 (2021) doi: 10.1038/s41467-021-26809-4.
Temmam, S. et al. Coronaviruses with a SARS-CoV-2-like receptor-binding domain allowing ACE2-mediated entry into human cells isolated from bats of Indochinese peninsula. Res. Sq. (2021) doi: 10.21203/RS.3.RS-871965/V1.
Wacharapluesadee, S. et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12, 972 (2021) doi: 10.1038/s41467-021-21240-1.