Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil
Authors
Paola Cristina Resende1*, João Felipe Bezerra2, Romero Henrique Teixeira de Vasconcelos2, Ighor Arantes3, Luciana Appolinario1, Ana Carolina Mendonça1, Anna Carolina Paixao1, Ana Carolina Duarte Rodrigues1, Thauane Silva1, Alice Sampaio Rocha1, Alex Pauvolid-Corrêa1,4, Fernando Couto Motta1, Dalane Loudal Florentino Teixeira5, Thiago Franco de Oliveira Carneiro5, Francisco Paulo Freire Neto6, Isabel Diniz Herbster7, Anderson Brandao Leite8, Irina Nastassja Riediger9, Maria do Carmo Debur9, Felipe Gomes Naveca10, Walquiria Almeida11, Mirian Livorati12, Gonzalo Bello3A, Marilda M Siqueira1A
A These authors share senior authorship
Affiliations
1 Laboratory of Respiratory Viruses and Measles (LVRS), Oswaldo Cruz Institute, Fiocruz
2 Universidade Federal da Paraíba (UFPB)
3 Laboratório de AIDS e Imunologia Molecular, Oswaldo Cruz Institute, Fiocruz.
4 Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, Texas, United States
5 Laboratório Central do Estado da Paraíba (LACEN-PB)
6 Institute of Tropical Medicine, Federal University of Rio Grande do Norte (UFRN)
7 Maternidade Escola Januario Cicco (MEJC-UFRN)
8 Laboratório Central do Estado do Alagoas (LACEN-AL)
9 Laboratório Central do Estado do Paraná (LACEN-PR)
10 Laboratório de Ecologia de Doenças Transmissíveis na Amazônia (EDTA), Leônidas e Maria Deane Institute, Fiocruz, on behalf of the COVIDNORTE team.
11 Secretary of Surveillance and Health of the Brazilian MoH (SVS-MS)
12 General Laboratories Coordination of the Brazilian Ministry of Health (CGLab/MoH)
Corresponding Author:
Paola Cristina Resende
[email protected]
Summary
On December 9th, the Brazilian Ministry of Health (MoH) recognised the first case of reinfection in the country. A 37-years-old healthcare worker resident in Northeast Brazil presented two clinical episodes of COVID-19 in June and October 2020, that were confirmed by RT-PCR in samples collected 116 days apart. Whole-genome sequencing revealed that the two infections were caused, respectively, by the two most prevalent SARS-CoV-2 Brazilian lineages B.1.1.33 (primo-infection) and B.1.1.28 (reinfection). Bayesian analysis indicates that the reinfection probably occurred between September 14th to October 11th, a few days before the second confirmed clinical episode of COVID-19, thus excluding the possibility of viral co-infection followed by long-term viral persistence. Notably, the B.1.1.28 virus detected at reinfection corresponds to a new emergent Brazilian viral lineage, initially detected in the Rio de Janeiro state, containing the mutation E484K in the Spike protein. We found this new viral variant, hereafter designated B.1.1.28(E484K), in several states from the South, Northeast, and North Brazilian regions and date its origin at August 27th, 2020 (July 14th - September 18th). These findings documented a classical SARS-CoV-2 reinfection case with the emerging Brazilian lineage B.1.1.28(E484K). Additionally, we provide evidence of this emerging Brazilian clade’s geographic dissemination outside the Rio de Janeiro state.
Keywords: COVID-19; SARS-CoV-2; reinfection; secondary infection.
Introduction
The efficiency and persistence of protective immunity naturally caused by the coronavirus disease (COVID-19) or vaccination are currently unknown. Some reinfection cases have been reported in different countries (1), but the differentiation between reinfection and viral persistence cases remains a challenge. The detection of two COVID-19 episodes months apart and caused by two different lineages of SARS-CoV-2 remains the most reliable evidence of a reinfection case. On December 9th, the Brazilian MoH recognised the first case of reinfection in the country (2). Here, we describe this classical reinfection case and highlight details about the genomic features of the two COVID-19 episodes. Additionally, we demonstrate that the second episode’s virus was related to the new viral variant, hereafter designated B.1.1.28 (E484K), circulating in several states from the South, Northeast, and North Brazilian regions.
Results
Case description
A 37-years-old woman without pre-existing comorbidities, healthcare worker (medical doctor) in Paraiba and resident in Rio Grande do Norte, neighbouring States in Brazilian Northeastern region, reported two clinical episodes of COVID-19. The first one in June and the second one in October 2020, comprising 116 days between the two episodes. On June 17th, the healthcare worker presented symptoms such as headache, runny nose, diarrhea, and myalgia, being classified as a mild COVID-19 case with no complications. The upper respiratory specimen (naso-oropharyngeal swabs - NPS) was collected from this patient six days later (June 23th). A second NPS was collected on 16th September as follow-up procedure. On October 11th, the patient presented new symptoms such as intense headache, ageusia, anosmia, and fatigue. This second infection was mild and evolved without complications as in the first episode, and a third NPS was collected two days later (October 13th).
This study was approved by the FIOCRUZ-IOC Ethics Committee (68118417.6.0000.5248 and CAAE 32333120.4.0000.5190) and the Brazilian Ministry of Health SISGEN (A1767C3). In addition, the patient reinfected with SARS-CoV-2 read and signed the free and informed consent form.
Diagnostic Laboratory findings
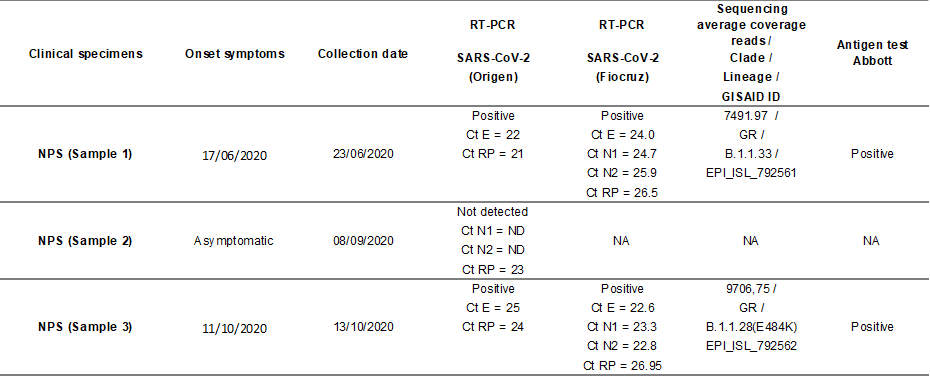
The three NPS samples collected were processed locally; the viral RNA was extracted and tested for SARS-CoV-2 using the real-time RT-PCR for the targets E (3) or N1 and N2 (4). After the diagnostic, Paraiba State Central Public Health Laboratory (LACEN-PB) sent the positive NPS and the serum specimens to the Respiratory Viruses and Measles Laboratory (LVRS) at Fiocruz (MoH National Reference Laboratory and WHO Reference Laboratory for Coronavirus) for confirmation and complementary analysis. Briefly, both positive samples were confirmed using the real-time RT-PCR protocol and the Ag-RDT Panbio COVID-19 Antigen Test (Abbott) directly from the clinical sample. According to the Brazilian MoH Technical Note, to confirm SARS-CoV-2 reinfection, two positive RT-PCR results separated by at least 90 days, independent of clinical conditions are necessary (5). The first and third clinical specimens collected on June 23th and October 13th, 2020, from the healthcare worker were positive for SARS-CoV-2 by real-time RT-PCR, while the second specimen collected on September 16th was negative. Both NPS positive specimens presented a high viral load, presumed by the low cycle threshold (Ct) values in the real time RT-PCR shown in Table 1.
Genomic findings
To distinguish between reinfection and long-term viral persistent hypothesis, we recovered the SARS-CoV-2 whole-genomes from the two positive NPS samples (EPI_ISL_792561 and EPI_ISL_792562) from the suspected reinfection case and 76 SARS-CoV-2 positive cases detected in the same state of Paraiba from April 6th to November 27th, 2020 (EPI_ISL_792563 to EPI_ISL_792638). The whole-genomes were recovered using Illumina or ONT Nanopore sequencing protocols previously established and used by Genomic Coronavirus Fiocruz Network to recover high-quality genomes (6,7). The FASTQ reads obtained were imported into the CLC Genomics Workbench version 20.0.4 (Qiagen A/S, Denmark), trimmed, and mapped against the reference sequence EPI_ISL_402124 available in EpiCoV database in the GISAID (https://www.gisaid.org/). The alignment was refined using the InDels and Structural Variants module, followed by the Local Realignment module and the final consensus obtained.
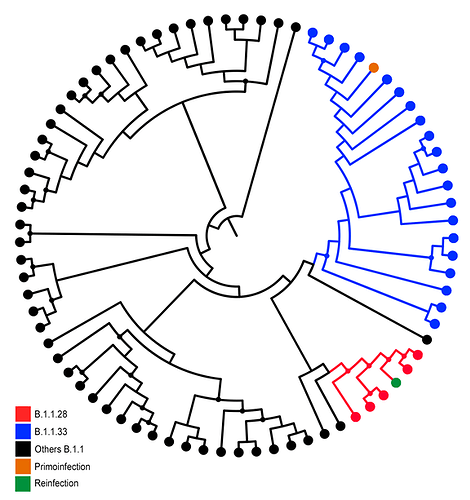
Maximum Likelihood (ML) phylogenetic analysis of all SARS-CoV-2 whole-genomes from Paraiba was conducted using PhyML8 and revealed two different viral lineages in the two COVID-19 episodes. In the first episode, the lineage B.1.1.33 was detected, whereas lineage B.1.1.28 was detected in the third (second positive) one (Figure 1), according to Pangolin lineage classification (9). These SARS-CoV-2 lineages were also detected in other samples from Paraiba state (Figure 1) and represent the most prevalent viral variants circulating in Brazil during the early epidemic phase (10,11). Of note, the B.1.1.28 sequences recovered from the reinfection case and from two additional cases in the Paraiba state harbor the E484K substitution (G23012A) in the Spike protein that was recently associated with a novel SARS-CoV-2 B.1.1.28 lineage circulating in Rio de Janeiro state (12).
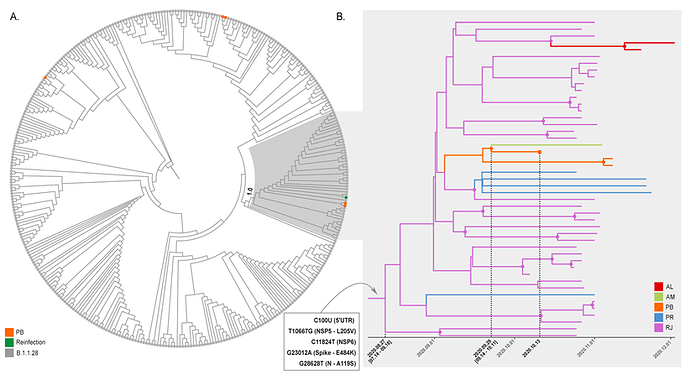
To better characterize the B.1.1.28 virus present in the second NPS positive sample of the reinfection case, we aligned it against all B.1.1.28 whole-genomes available in the EpiCoV database in GISAID by December 20th, 2020. Additionally, we also selected eight B.1.1.28 whole-genomes from Alagoas (n =2), Amazonas (n =1), and Parana (n = 5) states available at the Genomic Coronavirus Fiocruz Network database that also harbor the E484K substitution (EPI_ISL_792560, EPI_ISL_792639, EPI_ISL_792642, EPI_ISL_792645, EPI_ISL_792646, EPI_ISL_792650 to EPI_ISL_792652). The new ML phylogenetic tree revealed that the B.1.1.28 virus from the reinfection case branched in a highly supported (aLRT = 1) sub-clade with two sequences from Paraiba and 44 additional sequences recently sampled (October-December, 2020) in Brazil: 36 sequences from Rio de Janeiro recovered from the EpiCoV database in GISAID and the eight sequences from Alagoas, Parana, and Amazonas states that harbours the mutation S-E484K (Figure 2A). We identified five lineage-defining SNPs: C100U (5’UTR), T10667G (NSP5_L205V), C11824T (NSP6), G23012A (Spike_E484K), and G28628T (N_A119S) that distinguish sequences from the novel B.1.1.28 sub-clade from all other Brazilian B.1.1.28 sequences available. Because of the potential impact of the spike mutation E484K on viral phenotype (13-16), we designated this new emerging clade as B.1.1.28(E484K).
To further investigate the spatiotemporal emergence of the novel B.1.1.28(E484K) clade and the viral strain detected in the reinfection case, we conducted a Bayesian phylogeographic analysis of all 47 SARS-CoV-2 Brazilian sequences that branched within such clade. Time-scaled trees were estimated using a strict molecular clock model with a uniform substitution rate prior (8-10 x 10-4 substitutions/site/year), a GTR+I+G nucleotide substitution model, and the Bayesian skyline coalescent prior as implemented in BEAST 1.10 (17). Bayesian reconstructions traced the origin of the emerging B.1.1.28(E484K) lineage in the Rio de Janeiro State (PSP = 0.97) on August 27th (95% High Posterior Density [HPD]: July 14th – September 18th) and its subsequent dispersion from Rio de Janeiro to other States from the South (Parana) and Northeast (Alagoas and Paraiba) regions (Figure 2B). This phylogeographic reconstruction also supports a dissemination event from Paraiba State to Amazonas State and the branching of the B.1.1.28(E484K) sequence from the reinfection case with that from Amazonas with high support (PP = 1) (Figure 2B). The most recent common ancestor of B.1.1.28(E484K) sequences from the reinfection case and the Amazonas state was dated to September 29th (95% HPD: September 14th – October 11th), a few days earlier before the onset of reinfection symptoms (October 11th).
Discussion
We demonstrate that the first case of reinfection recognised by the Brazilian MoH on December 9th corresponds to a primoinfection with the Brazilian lineage B.1.1.33 and a reinfection with a novel emerging B.1.1.28 lineage harboring the mutation E484K in the Spike protein. The age of the common ancestor of the B.1.1.28(E484K) virus present in the second positive sample of the reinfection case and a non-related virus sampled in the Amazonas state provide a maximum limit for the reinfection episode between September 14th and October 11th, thus excluding the possibility of long-term persistence of the B.1.1.28(E484K) virus since primoinfection (before June 23rd, 2020).
Notably, the reinfection case reported here coincides with a recently reported case in the Bahia state that also described a primoinfection and reinfection with the B.1.1.33 and B.1.1.28(E484K) viral variants, respectively (18). These studies also confirm that the B.1.1.28(E484K) emerging variant initially described in the Rio de Janeiro state (12) is more widely distributed across different Brazilian states. Our analysis supports that the B.1.1.28(E484K) lineage probably emerged in the state of Rio de Janeiro around late August, but defining the precise location and time of emergence of this novel lineage will require a denser sampling from different Brazilian states during the second half of 2020.
The mutation E484K is located in the receptor-binding domain (RBD) and has also been recently described in an independent SARS-CoV-2 lineage rapidly spreading in South Africa (19). The rapid dissemination of these variants, combined with the ability of viruses harboring the mutation E484K to escape from neutralizing antibodies (13-16), and the recent cases of reinfection with the B.1.1.28(E484K) lineage detected in Brazil should raise concern about the potential impact of this mutation on infectivity, immune escape, and reinfection.
The findings presented here support that previous exposure to SARS-CoV-2 might not guarantee immunity in all cases. It reinforces the need to maintain the non-pharmacologic protective measures not only by individuals who tested negative, but also for those who already tested positive for SARS-CoV-2. Characterization of the immune response in individuals who presented reinfection with SARS-CoV-2 will be crucial to understand better the role of viral and host factors on this, apparently, rare phenotype.
Acknowledgment
We want to thank the funding support from CGLab/MoH (General Laboratories Coordination of Brazilian Ministry of Health), CVSLR/FIOCRUZ (Coordination of Health Surveillance and Reference Laboratories of Oswaldo Cruz Foundation), CNPq COVID-19 MCTI 402457/2020-0 and 403276/2020-9; INOVA Fiocruz VPPCB-005-FIO-20-2 and VPPCB-007-FIO-18-2-30; FAPERJ: E26/210.196/2020; FAPEAM (PCTI-EmergeSaude/AM call 005/2020 and Rede Genomica de Vigilancia em Saude - REGESAM). We also appreciate the support of all Genomic Coronavirus Fiocruz Network members, the Multi-user Research Facility of Biosafety Level 3 Platform of the Oswaldo Cruz Institute (IOC), Fiocruz, and the CGLab/MoH and Secretary of Surveillance and Health of the Brazilian MoH (SVS-MS). Additionally, we are thankful for all that contribute to the EpiCoV database from the GISAID initiative with high-quality SARS-CoV-2 genomes. All genomes used in this study are described in the acknowledgment Supplementary Table 1.
References
- Babiker A, Marvil C, Waggoner JJ, Collins M, Piantadosi A. The Importance and Challenges of Identifying SARS-CoV-2 Reinfections. J Clin Microbiol 2020.
- Brazilian Ministry of Health. Ministério da Saúde confirma primeiro caso de reinfecção por Covid-19. 10th December, 2020 2020. https://www.gov.br/saude/pt-br/assuntos/noticias/ministerio-da-saude-confirma-primeiro-caso-de-reinfeccao-por-covid-19.
- Biomanginhos FIOCRUZ. Kit Molecular SARS-CoV-2 (informações e consulta de manuais). 15th April, 2020 2020. https://www.bio.fiocruz.br/index.php/br/produtos/reativos/testes-moleculares/novo-coronavirus-sars-cov2.
- Prevention. CfDCa. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. 2020; (CDC-006-00019, Revision: 06): 80.
- Brazilian Ministry of Health Secretaria de Vigilância em Saúde. NOTA TÉCNICA Nº 52/2020-CGPNI/DEIDT/SVS/MS. In: Departamento de Imunização e Doenças Transmissíveis, Imunizações. C-GdPNd, editors. Brasilia, DF; 2020. p. 4.
- Resende PC, Motta FC, Roy S, et al. SARS-CoV-2 genomes recovered by long amplicon tiling multiplex approach using nanopore sequencing and applicable to other sequencing platforms. bioRxiv 2020.
- Nascimento VAD, Corado ALG, Nascimento FOD, et al. Genomic and phylogenetic characterisation of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Mem Inst Oswaldo Cruz 2020; 115: e200310.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59(3): 307-21.
- Rambaut A, Holmes EC, O’Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020.
- Resende P. C. D, E., Gräf T., Mir D., Motta F.C., Appolinario L., Paixão A. C., Mendonça A. C., Ogrzewalska M., Caetano B., Wallau G. L., Docena C., Santos M. C., Ferreirra J., Sousa Junior E., Silva S., Fernandes S., Vianna L. A., Souza L., Ferro J. F, Nardy V., Santos C., Riediger I., Debur M., Croda J., Oliveira, W, Abreu A, Bello G… Siqueira M. M. Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Frontier in Microbiology 2020.
- Candido DS, Claro IM, de Jesus JG, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 2020; 369(6508): 1255-60.
- Voloch CM, Silva F Rd, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv 2020.
- Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv 2021.
- Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020; 369(6506): 1014-8.
- Liu Z, VanBlargan LA, Rothlauf PW, et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv 2020.
- Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 2020; 9.
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 2018; 4(1): vey016.
- Vasques Nonaka CKMF, M.; Gräf, T.; Almeida Mendes, A.V.; Santana de Aguiar, R.; Giovanetti, M.; Solano de Freitas Souza, B. Genomic Evidence of a Sars-Cov-2 Reinfection Case With E484K Spike Mutation in Brazil. Preprints 2021.
- Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.
Figures and Tables
Table 1. Epidemiological and laboratorial data of the reinfection SARS-CoV-2 case detected in Northeast of Brazil.
NPS = naso-oropharyngeal swab; Ct = cycle threshold, E = envelope protein, N1 and N2 = nucleoprotein, RP = human RNAse P, NA = not applicable; ND = Not detectable.
Figure 01: ML of SARS-CoV-2 whole-genomes sequences from Paraiba. ML phylogenetic tree showing the branching pattern of SARS-CoV-2 whole genome sequences (29779 nt), from Paraiba (n = 77) classified within lineages B.1.1.28 (red), B1.1.33 (blue), and others B.1.1 (black). Sequences derived from the primo-infection and reinfection are both highlighted with different colors as indicate in the legend at left. Nodes with high statistical (aLRT ≥ 9.0) support are marked by circular shapes.
Figure 02: Emergence of the B.1.1.28(E484K) clade. A. ML phylogenetic tree of B.1.1.28 SARS-CoV-2 whole genome sequences (29779 nt) from Brazil (n = 376). Shaded box highlights the B.1.1.28(E484K) clade (n = 47) and its statistical support (aLRT = 1.0) is indicated in the cladogram. Sequences from Paraíba and the reinfection case are marked in orange and green, respectively. B. Time-scaled Bayesian MCC tree of SARS-CoV-2 whole genome sequences from the B.1.1.28(E484K) clade (n = 47). Branches are colored according to the most probable location state of their descendent nodes as indicated in the legend at right. The five lineage-defining SNPs are indicated at the MCCT root node. Circular shapes mark nodes with high statistical support (PP ≥ 9.0) and a square tip shape indicates the sequence from reinfection case.
Supplementary Materials
Supplementary Table 1. GISAID acknowledgment table Suppelemtary Table 1. gisaid_hcov-19_acknowledgement_table_2021_01_10_16.pdf (32.3 KB)