Localized Sequence Identities Between SARS-CoV-2 Spike Protein and Lima Bean Agglutinin Specific for ABO Blood Group A
William R. Gallaher
Mockingbird Nature Research Group, Pearl River LA 70452 and
Department of Microbiology, Immunology and Parasitology, School of Medicine, Louisiana State University Health, New Orleans LA 70112
Abstract: A serendipitous alignment, of the N-terminal domain of SARS-CoV-2 spike protein with the lima bean lectin specific for ABO blood group A, supports sequence and structural homology of these proteins. In a possible example of molecular mimicry, this homology defines a potential binding groove for ABO blood group A glycoside on the outer surface of the N-terminal domain of the spike protein of SARS-CoV-2. Such binding, if confirmed experimentally, may identify a second receptor binding site on the spike protein that may, in turn, correlate with the recently reported differential severity of COVID-19 between patients of different ABO blood groups.
Evidence has been accumulating over the last few months correlating risk of severe infection due to SARS-CoV-2 to ABO blood type (1,2). Specifically, in a large clinical cohort, Ellinghaus et al. (3) found a 1.45 elevated risk for individuals with blood group A vs. a reduced risk of 0.67 for individuals with blood group O. A similar correlation had been previously observed for SARS-CoV in 2005 (4). A mechanism for this consistently observed effect in several countries with diverse populations has not been demonstrated.
An interaction between SARS-CoV-2 and human blood groups is consistent with numerous studies documenting interactions between other coronaviruses and a variety of glycosides (reviewed in 5), especially sialic acid as the terminal sugar in a number of different glycopeptides and glycolipids. These interactions have been localized to the S protein, and more specifically to the N-terminal of two readily distinguishable domains in the S1 attachment subunit, frequently designated NTD or S1A. For bovine coronavirus, MERS and the JHM strain of mouse hepatitis virus, the S1A domain has been found to bear significant structural similarity to the galectin family of lectins, and to bind sialic acid. The second domain, designated S1B, has been found to bind protein receptors such as the ACE2 receptor in the case of SARS-CoV of 2003 and SARS-CoV-2. Importantly, for neither of the latter two viruses has any function been associated with the NTD (S1A) domain, which in SARS-CoV-2 accounts for 291 amino acids, nearly half of the S1 subunit, and 23% of the entire spike protein.
Several plant lectins have been identified that are specific for the human ABO blood group glycosides, and have been useful for blood typing for many decades (6). An ABO blood group A-specific lectin is lima bean agglutinin (LBA), which selectively binds the terminal N-acetyl-galactosamine of the group A polysaccharide.
The S1A domains of SARS-CoV and SARS-CoV-2 are structurally similar and readily alignable. They are also structurally similar to that of other coronaviruses, especially MERS, in spite of a great deal of sequence diversity that challenges any alignment with high confidence. Nevertheless, I decided to test the hypothesis that the NTD of SARS-CoV-2 may be identifiable as a galectin capable of binding human blood group glycosides.
I compared the protein sequences of the S1 glycoprotein of SARS-CoV-2 (NC_045512) and LBA (Genbank AJ271874)using Clustal X 2.1 (7), on the off chance that some similarity could be discerned. Indeed, only a small fraction of the proteins showed sequence identity or a high degree of similarity. However, there were three regions with multiple identities or high similarities, as shown in Table 1:
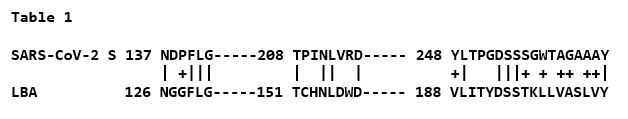
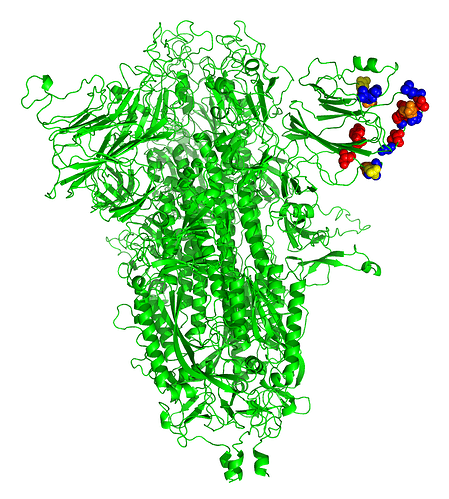
While this degree of similarity is not very impressive entirely on its own, superimposing the identities on the known x-ray structure of the NTD produced a rather more notable result, as shown in Figure 1.
FIGURE 1. Superimposition of amino acid identities between SARS-CoV-2 spike protein and LBA. The SARS-CoV-2 spike model was created in SWISS-MODEL (8) by replacing the modifications in the sequence made by Walls et al (9) to stabilize the protein for crystallography (pbd 6VXX). The model was rendered in PyMOL (v1.8.0.6), an open source molecular visualization system created by Warren DeLano that is commercialized by Schrodinger, Inc. (New York City, New York; https://pymol.org/2/ ). Upon this model were superimposed the amino acid identities between SARS-CoV-2 and LBA, plus two highly similar amino acids (G/P; S/T) adjacent to the two identical tripeptides FLG and DSS in both sequences. Color code: Red, hydroxylated, S, T, Y; orange, carboxylated, D; yellow, amidated, N; blue, framework, F, L and G.
The identical amino acids between the two proteins not only cluster at the outer end of the beta-sheeted NTD structure, but are aligned on either side of an apparent groove in the surface of the domain. The first region NDPFLG defines framework residues underlying the groove at upper left, The second region TPINLVRD is at lower left at the surface, and the third, longest, region YLTPGDSSSGWTAGAAAY defines the right outermost region of identity. As the brighter color residues indicate, the groove is enriched in carboxylated (D), hydroxylated (S,T,Y) and amidated (N) residues that are common in binding sites for carbohydrates through hydrogen bonding.
The clear implication of this sequence and structural alignment, between the NTD of SARS-Cov-V-2 spike protein and an ABO group A specific lectin, is that the delineated groove may define an ABO blood group binding groove on the surface of SARS-CoV-2. Such a groove specific for type A glycosides would be consistent with the differential risk patterns for serious COVID-19 observed globally.
The NTD is highly glycosylated, creating the possibility of steric hindrance of binding a ligand in this area. However, none of the predicted glycans directly overlap these outermost polar residues.
While these identities are not seen with SARS-CoV of 2003, that sequence can be easily aligned with SARS-CoV-2, raising the possibility that the NTD of SARS-CoV of 2003 may also have similar lectin-like characteristics.
As mentioned above, MERS spike protein has been shown to bind sialic acid, with binding associated with the NTD of S1, confirming its lectin-like activity (10). Recently, Qing et al (5) have extended this to identify an amino acid residue, 222N, that, when mutated to D, significantly abrogates binding of sialic acid in the case of MERS virus. While MERS (Genbank NC_ 019843) and SARS-CoV-2 are highly divergent in their NTD sequences, Clustal X 2.1 aligns the following region, shown in Table 2, with two identities and seven high similarities.

The identical 226 N of MERS and 211 N in SARS-CoV-2 aligns with the 154 N of LBA. The highly similar 230 E of MERS and 215 D of SARS-CoV-2 also aligns with 158 D of LBA.
These similarities strain the bounds of coincidence, at the least. While there is no indication that SARS-CoV-2 binds sialic acid at all, this regional overlap of amino acid sequences involved in glycoside binding is very suggestive that the NTD of SARS-CoV-2 in this distal end of the protein domain may provide a second, hitherto unidentified, receptor function for SARS-CoV-2, quite possibly binding the ABO blood group glycoside.
SARS-CoV-2, and potentially SARS-CoV of 2003, may well be added to a lengthening list of coronaviruses that utilize both S1A and S1B as receptor binding domains, the first as a lectin for relatively low affinity initial binding, and then, higher affinity binding of the second to a protein receptor such as the ACE2 receptor defined for both viruses. In the case of SARS-CoV-2, this second receptor may indeed be the ABO blood group A-specific ligand, N-acetyl-galactosamine.
The potential of targeting ABO blood group antigens for therapy against these viruses has already been suggested (11,12). The possible identification of a second receptor for SARS-CoV-2 for the ABO blood group A determinant heightens this potential in ways difficult to predict. However, until definitive molecular identification of a second receptor and its ligand are defined experimentally, it would be extremely ill-advised to attempt to translate these possibilities into interventional treatments. Much experimentation lies ahead to define the function of the NTD (S1A) domain and determine whether firmer knowledge of that function may lead to antiviral treatments.
What is defined here is merely a hypothesis derived from very limited, but intriguing, sequence and structural homologies, no more. I would respectfully submit, however, that this admittedly serendipitous alignment is more than we knew yesterday, and potentially of some significance moving forward.
Acknowledgments: The author is a long-retired Professor Emeritus, working from a home-based consulting office, with no outside or institutional support. However. I am greatly indebted and grateful for my longtime friend and collaborator of over 30 years, Dr. Robert F. Garry of the Tulane School of Medicine, New Orleans, who graciously and tirelessly provides me with extensive graphic assistance to complement my limited computational and graphic capabilities. I also thank my son, MSGT Andrew D. Gallaher USMC, for help in editing the manyscript.
References:
- Zhao J et al. (2020) Relationship between the ABO blood group and the COVID-19 susceptibility. March 27; https://www.medrxiv.org/content/10.1101/2020.03.11.20031096v2
- Zietz M, Tatonetti NP. (2020) Testing the association between blood type and COVID-19 infection, intubation, and death. April 11; https://www.medrxiv.org/content/10.1101/2020.04.08.20058073v1.
- Ellinghaus D et al (2020) Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med June 17; DOI: 10.1056/NEJMoa2020283
- Cheng Y et al. (2005) ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA 293:1450-1451.
- Qing E, Hantak M, Perlman S, Gallagher T. 2020. Distinct roles for sialoside and protein receptors in coronavirus infection. mBio 11:e02764-19. https://doi.org/10.1128/ mBio.02764-19.
- Sparvoli F et al (2001) Lectin and lectin-related proteins in lima bean (Phaseolus lunatus L.) seeds: biochemical and evolutionary studies. Plant Mol Biol 45: 587-597
- Larkin MA et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947-2948
- Waterhouse, A et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296-W303
- Walls AC et al (2020) Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181:281-292. https://doi.org/10.1016/j.cell.2020.02.058
- Li W et al (2017) Identification of Sia-binding by MERS-CoV spike glycoprotein. Proc Nat Acad Sci USA: E8508-E8517; DOI: 10.1073/pnas.1712592114
- Breiman A, Ruvën-Clouet N, Le Pendu J. (2020)Harnessing the natural anti-glycan immune response to limit the transmission of enveloped viruses such as SARS-CoV-2. PLoS Pathog 2020;16(5):e1008556-e1008556
- Guillon P et al (2008) Inhibition of the interaction beteen the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. 2008;18:1085–93.