It has become clear that there is a wide spectrum of illness and individual response to infection by nCoV2019, from very mild illness to induction of a cytokine storm and death. Some of this variation may lie in the fact that human infection always occurs against the backdrop of our “virome”. This includes the expression of endogenous viruses, that comprise 8% of the human genome, as well as any recent experience with other viral agents.
The spike protein belongs to Class I of fusion.entry proteins. These are constructed of a series of structural and functional domains and motifs that may be highly conserved in localized regions. Insofar as sequence motifs of nCoV2019 show high similarity to the Class I fusion/entry proteins of endogenous viruses expressed as part of the human genome – to which we would be expected to be tolerant – response to nCoV2019 could be significantly affected.
Indeed, alignment of the endogenous elements Syn1 found on human chromosome 7, or Syn2 found on chromosome 6, or HERV-K expressed from chromosome 6, all show a number of sequence motifs with significant similarity to nCoV2019 spike protein.
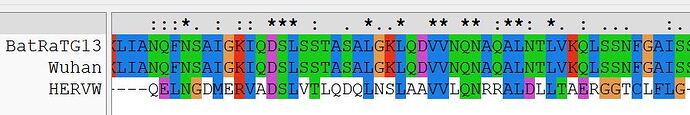
The first region comprises the majority of Wuhan HR1a of S2, with the corresponding region of HERV-W (Syn1). Basically, the alignment shows that HR1a of nCoV2019 and HR1 of Syn1 are directly related to one another.
Since HR2 nCoV2019 very obviously corresponds to retroviral HR2, this leaves HR1b, the central helix of nCoV2019, as the missing element when comparing coronaviruses to the other virus families with Class I fusion/entry proteins, such as the retroviruses, filoviruses and arenaviruses.
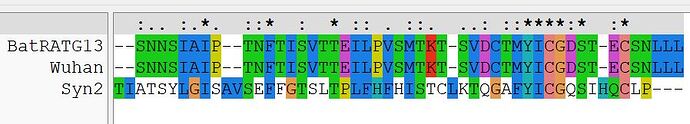
A second region of nCoV2019 that is of unknown function is the amino terminal region of S2, that in Wuhan follows the novel RRAR furin cleavage site I first disclosed here in January. This region of nCoV2019 also convincingly aligns with both Syn2 as well as HERV-K.
Shown first is the alignment with Syn2, with an extraordinary, nearly identical hexapeptide in each.
Over the same region, there is also an extraordinary similarity with HERV-K.
Note that identical amino acids tend to be in every other position. In a beta sheet, these would lie on one side of the sheet, rendering its stereochemistry from that aspect identical, as shown in the following figure.
Within the boxed regions, stereochemistry is identical. Any element of the host response that has seen the one in HERV-K would be expected to respond identically to that within nCoV2019.
Given the high similarity of this peptide region in nCoV2019 to endogenous retroviral peptide sequence, one hypothesis of the purpose of this region in nCoV2019 would be as a toleragen, or, when clipped out of the protein between the two endoproteolytic sites in S, as a soluble decoy peptide.
There is another region of similarity within the receptor binding domain of nCoV2019 S1, to HERV-W, likewise expected to induce tolerance.
Finally, as we disclosed here on January 30 in the pdf uploaded for free download, there is in nCoV2019 S1 a peptide region with the critical amino acids characteristic of retroviral immunosuppressive peptides. The alignment I showed then is repeated here.

While this peptide has not been shown to actually have immunosuppressive properties, it is conserved in both nCoV2019 and RaTg1. It should be noted that one of the characteristics shown by patients with severe nCoV2019 disease is lymphpenia, indicative of an immunosuppressed state.
There are therefore a number of peptide regions in nCoV2019 having significant overlap with sequences expressed by human endogenous viruses, or derived therefrom during viral evolution. These could have significant effects on patient response to nCoV2019 infection, including both tolerance and immunosuppression.
Finally, antecedent infection with any number of human viruses can render a patient temporarily refractory to the induction of interferon and other modulators of the human non-specific or specific immune response. The virome of an individual may be a factor in understanding the wide spectrum of host response to nCoV2019 infection.
Bill Gallaher