Repeated loss of ORF8 expression in circulating SARS-CoV-2 lineages
Ryan Hisner1, Federico Gueli2, Thomas P. Peacock3*
1Independent Researcher, USA
2Independent Researcher, Italy
3Department of Infectious Disease, Imperial College London, UK
*Corresponding author – [email protected]
Introduction
SARS-CoV-2, like other coronaviruses, expresses a variety of non-structural, structural and accessory proteins. Accessory proteins in coronaviruses are defined as proteins that are not essential for virus growth in cell culture. Accessory proteins may have important roles in vivo and are often involved in modulation of host cell pathways such as the interferon suppression, cellular immunity, apoptosis and autophagy.
The ORF8 protein of SARS-CoV-2 lies within the least conserved region of the genome between SARS-CoV and SARS-CoV-2[1]. SARS-CoV-2 ORF8 encodes a ~121aa accessory protein that is thought to be secreted via an N-terminal signal peptide[2, 3]. The exact role and the importance of ORF8 remains contentious[4]. Several studies have found ORF8 has only minimal impact on pathogenicity or infectivity in a mouse model[5, 6]. Suggested roles of ORF8 including a virokine (a viral cytokine mimic)[7], an inhibitor of host MHC expression[8, 9], a protein with histone-like activity[10], a modulator of interferon[11], a inducer of endoplasmic reticulum stress responses[12], and an inducer of inflammation[13]. Some of these activities have been shown to still be enacted by virus strains lacking ORF8 expression, suggesting a high level of redundancy between SARS-CoV-2 proteins[9].
SARS-CoV-2 variants are most well known for having large numbers of mutations in their spike proteins that alter virus pathogenicity, transmissibility and antigenicity[14]. However, a number of variants have mutations throughout other regions of the genome, including in accessory proteins or in regulatory RNA sequences. The ORF8 region, in particular, has been a hotspot for mutations, particularly in later Omicron lineages.
Here we describe how currently circulating and growing SARS-CoV-2 lineages have convergently reduced or removed ORF8 expression, leading to a situation where we predict that soon nearly all circulating lineages will either poorly express, or completely lack ORF8 expression.
BA.5 and XBC contain mutations in their ORF8 transcriptional regulatory sequences (TRS) that are predicted to reduce or ablate ORF8 expression
Expression of all coronavirus genes (other than ORF1ab) require the virus to generate subgenomic mRNAs (sgmRNAs). Generation of sgmRNAs requires conserved transcriptional regulatory sequences (TRSs) which are found upstream of all ORFs in the coronaviral genome. During synthesis of the negative-sense copy of the genome, upon reaching the structured TRS sequences, the coronaviral polymerase can stall, drop off its template and reattach at the TRS-leader (TRS-L) sequence at the 5’ end of the template. This generates a chimeric viral RNA, which when copied into the positive sense, acts as a specific mRNA for the immediate downstream gene. De novo, or enhanced homology between the body TRS sequences (TRS-B) and the TRS-L can lead to de novo or enhanced expression of that specific TRS[15]. Similarly, disruption of the core TRS-B sequences likely results in ablated or greatly reduced expression.
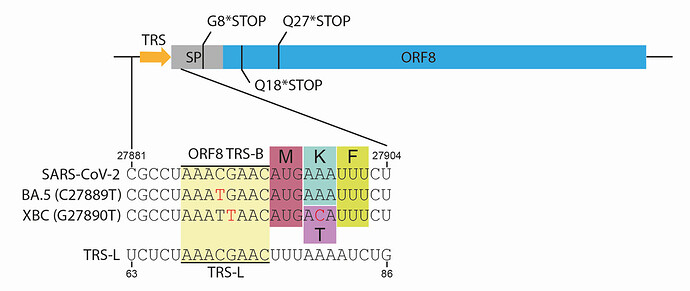
Figure 1. Schematic of ORF8 gene showing the most common truncations and TRS mutations. *STOP indicates stop codons, SP = signal peptide, TRS = transcriptional regulation site, TRS-B = TRS body, TRS-L = TRS leader. Red text indicates mutations compared to reference sequence.
A number of recent SARS-CoV-2 lineages contain mutations that reduce the homology between the ORF8 TRS-B (which is found immediately upstream of the ORF8 open reading frame). This includes within the core TRS sequence (Figure 1), which is likely to have the most dramatic effect. The most widespread of which is BA.5, for which the mutation C27889T which lies in the middle of the core TRS, is found in nearly all sequences. XBC is a complex recombinant between BA.2 and Delta[16, 17], XBC.1.6 is a sublineage of XBC with further antigenic mutations that is continuing to grow in several countries. All XBC sequences and sublineages contain G27890T, another core ORF8 TRS mutation (Figure 1). Both C27889T and G27890T are very likely to either reduce or entirely ablate ORF8 sgmRNA expression, although to what extent requires experimental validation.
Most XBB.1 sublineages have truncated ORF8 proteins
The current most abundant and fast-growing examples of lineages containing ablated ORF8 expression are sublineages of XBB.1 (Figure 2), including XBB.1.5, XBB.1.9, and XBB.1.16. All these lineages contain an ORF8 protein with a stop codon at position 8 (ORF8:G8*STOP) which lead to a protein truncated before the end of the signal peptide and therefore extremely unlikely to maintain any ORF8 function. This truncation was likely inherited from a common XBB.1 ancestor, a lineage which itself largely contains this truncation (although it is not a lineage-defining mutation).
The fastest growing, and most abundant SARS-CoV-2 lineages are predicted to have lost ORF8 expression.
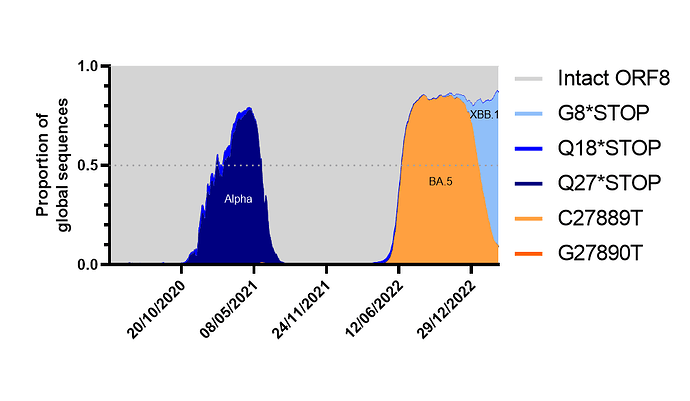
Figure 2. ORF8 expression over time as a proportion of total global sequences (from April 2020 to to 18th April 2023, as of 26th April 2023). Analysis made using cov-spectrum.org with open data. Lineages making up the highest proportion of waves annotated onto graph.
We calculated the proportion of sequences that had either fully expressed, full length ORF8 (based on a lack of stop codons or ORF8 TRS-B mutations) or the most common truncations or TRS ablations and plotted them over time. Other than a drop during the Alpha period (which is likely to be heavily biased due to a predominance of European sequences at the time and a lack of travel) we found that the BA.5 era resulted in the first major drop in predicted ORF8 expression, this was followed by a period of massive diversification of BA.2 and BA.5 lineages eventually resulting in XBB.1 sublineages winning out and causing a further rapid and dramatic drop[16]. Assuming no new fast-growing lineages emerge from a non-XBB.1, non-BA.5 background (ie from BA.2.75 which is the largest remaining family of lineages that still contain an intact ORF8) its likely by June 2023 less than 10% of circulating lineages will have an intact ORF8.
Concluding remarks
As viruses adapt to a new host it is often seen that selection can occur at different speeds, sometimes rapidly, for example with D614G in SARS-CoV-2, sometimes much more slowly due either to complicated combinations of compensatory mutations required, or due to adaptations being fine tuning. Loss or ablation of ORF8 is not restricted to recent sequences - one early cluster of SARS-CoV-2 in Singapore contained a large deletion which resulted in complete loss of ORF7b and ORF8[18]. Furthermore, the early variant of concern (VOC) Alpha as well as several other less abundant examples had ORF8 truncations, or mutations in the upstream TRS (For a complete list see Table 1) [19, 20]. Clearly ORF8 is not essential for efficient transmission and replication of SARS-CoV-2, and recent growth of lineages that do not express ORF8 by several alternative mechanisms suggests it may even be advantageous in certain scenarios to ablate its expression entirely (although we cannot rule out these mutations could be neutral genetic hitchhikers).
We are currently heading for a situation where most, if not all, circulating SARS-CoV-2 lineages lack ORF8 expression. However, it is entirely possible re-emergence of a fit lineage derived from, or a recombinant of a waning lineage like BA.2.75, XBB.2, or XAY could occur and could again result in circulating lineages containing ORF8. Alternatively, all the contemporary mechanisms of ORF8 ablation are reversible with a single nucleotide change suggesting spontaneous reversion of these ablations could occur easily.
Further work should attempt to quantify the impact of the different TRS mutations on ORF8 sgmRNA and protein expression. Furthermore, it would be informative to generate contemporary virus isolates (such as in a XBB.1.5 genetic background) to measure isogenic changes in ORF8 expression (ie with or without the truncation) and characterise the impact of this deletion in vitro and in vivo, although we cannot exclude that ORF8 may have a highly species specific role that is obscured when using rodent models.
| Pango Lineage | ORF8 ablation | Predicted effect |
|---|---|---|
| Pre-Omicron lineages | ||
| B.1.1.7(Alpha) | Q27*STOP | Truncation |
| B.1.1.318 | Δ27887-27901 | Removal of entire ORF8 TRS, in-frame fusion between ORF7b and ORF8 |
| B.1.429 (aka part of Epsilon) | G27890T | Suboptimal core TRS, reduced/ablated expression |
| B.1.640 | Q27*STOP | Truncation |
| P.1.8 | G8*STOP | Truncation |
| B.1.625 | Δ27897-27901 | Out of frame deletion leading to premature stop codon, expression of 18aa peptide |
| B.1.630 | C27889T | Suboptimal core TRS, reduced/ablated expression |
| Post-Omicron lineages | ||
| BA.5 | C27889T | Suboptimal core TRS, reduced/ablated expression |
| BA.2.65 | Q18*STOP | Truncation |
| Some BA.2.12.1 (inc BG.2) | Q18*STOP | Truncation |
| XBC | G27890T | Suboptimal core TRS, reduced/ablated expression |
| Most XBB.1 | G8*STOP | Truncation |
Table 1. Lineages with predicted ORF8 loss
Acknowledgements
The authors would like to thank Josette Schoenmakers, Nicholas Frohberg, Hitoshi Sakaguchi, @wanderer_jasnah, and Darren Martin for their useful discussions and feedback. The authors would particularly like to thank Chaoran Chen and all the developers behind covSPECTRUM for making this analysis possible[21]. Finally, the authors would like to thank all data collectors and submitters who provide the underlying data for open.covSPECTRUM.
References
-
Tan, Y., et al., Novel Immunoglobulin Domain Proteins Provide Insights into Evolution and Pathogenesis of SARS-CoV-2-Related Viruses. mBio, 2020. 11(3): p. e00760-20.
-
Matsuoka, K., et al., SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer. Journal of Biological Chemistry, 2022. 298(3).
-
Flower, T.G., et al., Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proceedings of the National Academy of Sciences, 2021. 118(2): p. e2021785118.
-
Zinzula, L., Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem Biophys Res Commun, 2021. 538: p. 116-124.
-
Silvas, J.A., et al., Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice. Journal of Virology, 2021. 95(17): p. e00402-21.
-
McGrath, M.E., et al., SARS-CoV-2 variant spike and accessory gene mutations alter pathogenesis. Proceedings of the National Academy of Sciences, 2022. 119(37): p. e2204717119.
-
Kriplani, N., et al., Secreted SARS-CoV-2 ORF8 modulates the cytokine expression profile of human macrophages. bioRxiv, 2021: p. 2021.08.13.456266.
-
Zhang, Y., et al., The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-I. Proceedings of the National Academy of Sciences, 2021. 118(23): p. e2024202118.
-
Moriyama, M., et al., Enhanced inhibition of MHC-I expression by SARS-CoV-2 Omicron subvariants. Proceedings of the National Academy of Sciences, 2023. 120(16): p. e2221652120.
-
Kee, J., et al., SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature, 2022. 610(7931): p. 381-388.
-
Chen, J., et al., Severe Acute Respiratory Syndrome Coronavirus 2 ORF8 Protein Inhibits Type I Interferon Production by Targeting HSP90B1 Signaling. Front Cell Infect Microbiol, 2022. 12: p. 899546.
-
Wang, X., et al., SARS-CoV-2 ORF8 Protein Induces Endoplasmic Reticulum Stress-like Responses and Facilitates Virus Replication by Triggering Calnexin: an Unbiased Study. Journal of Virology, 2023. 97(3): p. e00011-23.
-
Lin, X., et al., ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience, 2021. 24(4).
-
Carabelli, A.M., et al., SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nature Reviews Microbiology, 2023: p. 1-16.
-
Mears, H.V., et al., Emergence of new subgenomic mRNAs in SARS-CoV-2. bioRxiv, 2022: p. 2022.04.20.488895.
-
Roemer, C., et al., SARS-CoV-2 Evolution, Post-Omicron. virological. org. 911.
-
Pangilinan, E.A.R., et al., Analysis of SARS-CoV-2 Recombinant Lineages XBC and XBC.1 in the Philippines and Evidence for Delta-Omicron Co-infection as a Potential Origin. bioRxiv, 2023: p. 2023.04.12.534029.
-
Su, Y.C.F., et al., Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio, 2020. 11(4).
-
Rambaut, A., et al., Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org, 2020.
-
Peacock, T.P., et al., SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J Gen Virol, 2021. 102(4).
-
Chen, C., et al., CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics, 2021. 38(6): p. 1735-1737.