Wolfgang Maier1, Simon Bray1, Marius van den Beek2, Dave Bouvier2, Nathaniel Coraor2, Milad Miladi1, Babita Singh3, Jordi Rambla De Argila3, Dannon Baker4, Nathan Roach5, Simon Gladman6, Frederik Coppens7,8, Darren P Martin9, Andrew Lonie6, Björn Grüning1, Sergei L. Kosakovsky Pond10, and Anton Nekrutenko2
1 University of Freiburg / usegalaxy.eu, Freiburg, Germany
2 The Pennsylvania State University / usegalaxy.org, University Park, PA, USA
3 GalaxyWorks Inc, Baltimore, MD, USA
4 Centre for Genomic Regulation, Viral Beacon Project, Barcelona, Spain
5 Johns Hopkins University / usegalaxy.org, Baltimore, MD, USA
6 University of Melbourne / usegalaxy.org.au, Melbourne, Australia
7 Ghent University, Ghent / usegalaxy.be, Belgium
8 VIB Center for Plant Systems Biology, Ghent, Belgium
9 University of Cape Town, Cape Town, South Africa
10 Temple University, Philadelphia / datamonkey.org, PA, USA
Why?
Global inequality in COVID-19 response is not limited to vaccine development and distribution. Virtually no one talks about another type of inequality—inequality at the research level and specifically at the level of data analysis. We continuously hear about UK, South African, and Brazilian “variants of concern”. This is not because there is something peculiar about these countries. Rather, it is indicative of the monitoring infrastructures that they possess. Surely, there are other “variants of concern”. We simply do not know about them as countries where they originate do not have means for reliable monitoring and early detection.
Furthermore, even in developed countries different research groups perform sequence analyses in distinct ways with some approaches being more appropriate than others. This multitude of analytical approaches makes it hard to integrate newly acquired knowledge and compare results across studies.
Our work is the direct response to these challenges. We assembled a global data analysis consortium leveraging curing edge public computational infrastructure from the US, Europe, and Australia to deliver a platform that can be used now by any researcher from any country. Our workflows can be used to analyze existing or new data no matter how big or complex these datasets may be. We also emphasize the fact that the access to raw data is essential and demonstrate how sequencing read data can be used to uncover potentially important viral heterogeneity within samples.
What do we want from Virological?
We need your help in testing and fine tuning the workflows, documentation, and tutorials! Our system is housed on three globally distributed public Galaxy instances in the US (http://usegalaxy.org), the EU (http://usegalaxy.eu), and Australia (http://usegalaxy.org.au). Each is capable of supporting thousands of users running hundreds of thousands of analyses per month. Anyone can create an account and obtain immediate access to as much computation as one might reasonably need (with a limit on the number of concurrent analyses) and 250 Gb of disk space, which can be increased based on the needs of an individual user.
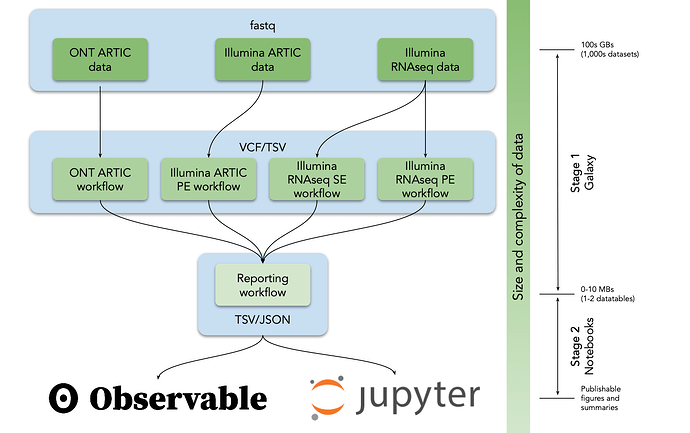
We currently have five workflows for dealing with Ampliconic data generated with Illumina and ONT as well as RNAseq (WGS) data:
How do I use it?
We created a documentation page at Global platform | COVID-19 analysis on usegalaxy.★
Where do I get help?
You can either email us ([email protected]) or use our help system at https://help.galaxyproject.org