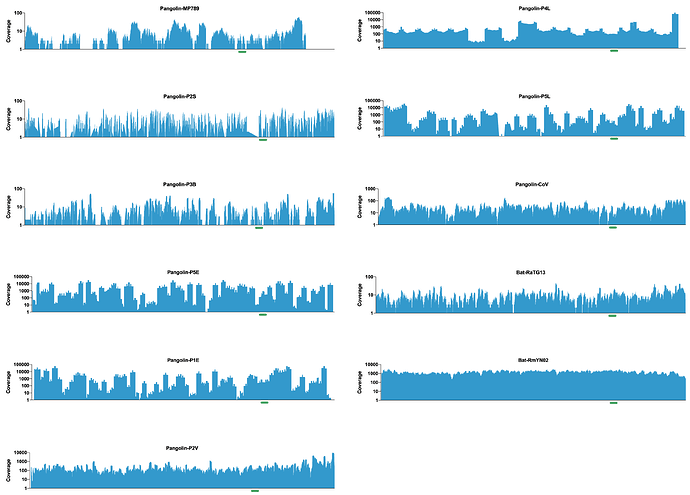
To further investigate the authenticity of recent pangolin and bat SARS-like coronaviruses, I downloaded all the raw data and assembled all the genomes using bwa mem. All the genomes assembled according to what had been described in the various papers and important features like the receptor binding domains were fully resolved (see figure - RBD highlighted with a green bar under the coverage plot).
Samples
| Name | Reference | Type | Study | PMID | BioProject | reads |
|---|---|---|---|---|---|---|
| MP789 | EPI_ISL_412860 | pangolin | Li et al | 31652964 | PRJNA573298 | SRR10168377 SRR10168378 |
| Pangolin-CoV | EPI_ISL_410721 | pangolin | Xiao et al | 32380510 | PRJNA607174 | SRR11119759 SRR11119762 SRR11119765 SRR11119766 SRR11119767 SRR12053850 |
| RmYN02 | EPI_ISL_412977 | bat | Zhou et al | 32416074 | PRJNA656060 | SRR12432009 SRR12464727 |
| RmYN01 | EPI_ISL_412976 | bat | Zhou et al | 32416074 | NMDC1001304 | did not download because of slow speeds |
| P1E | EPI_ISL_410539 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093266 |
| P2S | EPI_ISL_410544 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093265 |
| P2V | EPI_ISL_410542 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093271 |
| P3B | EPI_ISL_410543 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093270 |
| P4L | EPI_ISL_410538 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093269 |
| P5E | EPI_ISL_410541 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093268 |
| P5L | EPI_ISL_410540 | pangolin | Lam et al | 32218527 | PRJNA606875 | SRR11093267 |
| RaTG13 | EPI_ISL_402131 | bat | Zhou et al | 32015507 | PRJNA606165 | SRR11085797 SRR11806578 |
Data
Fastq files for each sample were downloaded directly from ENA as single-read data. Consensus genomes were downloaded directly from NCBI and used as reference sequences for genome assembly.
Methods
Sequencing data was uncompressed and aligned in single-read mode to each relevant reference genome using bwa mem with default settings and saved as an aligned bam file using samtools:
gunzip -cd {input_reads.gz} | bwa mem -t 8 {reference.fasta} /dev/stdin | samtools view -q 1 > {output.bam}
Data
All relevant data - including assembled bam files and high resolution coverage plots - can be downloaded from our Google Cloud repo and via our project page.