The African Centre of Excellence for the Genomics of Infectious Diseases [ACEGID], Redeemer’s University, Ede, Nigeria [RUN], and the Nigeria Centre for Disease Control [NCDC] report twenty-four [24] additional genome sequences of SARS-CoV-2 from Nigeria. These sequences are available at: [GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria].
As a member of the Molecular Laboratory Network of the Nigeria Centre for Disease Control [NCDC], clinical specimens [specifically saliva, nasopharyngeal and nasal swabs] from suspected COVID-19 cases were sent to ACEGID, Redeemer’s University, for confirmatory testing, sequencing and molecular characterization. Viral RNA was extracted using the QiAmp viral RNA mini kit [Qiagen]. RT-qPCR was carried out using the DAAN RT-qPCR assay which confirmed the presence of SARS-CoV-2 viral RNA. Metagenomic sequencing libraries were prepared from total RNA as we previously described [Matranga et al ., 2016], and sequenced using the two Illumina MiSeqs in the sequencing platform of ACEGID.
Genome Assembly and Quality control
We carried out genome assembly using our publicly available software [viral-ngs v2.0] implemented on the DNA nexus cloud-based platform. We assembled 24 genomes [18 full and 6 partials]. We carried out quality control using fastqc [Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data].
Phylogenetics and Lineage Delineation
Representative HCoV whole genome sequences of each of the seven lineages circulating in Nigeria were obtained from GISAID and aligned with all full genomes from Nigeria so far. The sequences were aligned using MAFFT v7.310 [Katoh et al., 2009] and tree reconstruction using FastTree v2.1.11 [Price et al., 2009].
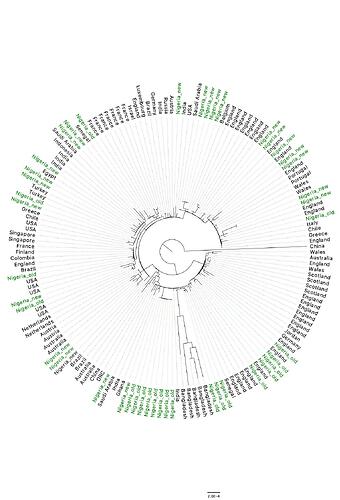
Four of our new sequences clustered closely together and formed a separate clade which strongly suggests local community transmission [Figure 1]. Epidemiological data confirmed that all four sequences are from patients in a community in Ekiti State, Nigeria with three of these four patients being members of the same family. We also observed a couple other new sequences clustering closely together, further investigation however revealed these sequences are from follow-up samples obtained from the same patients.
Using Pangolin software [Rambaut et al ., 2020], we assigned the sequences to global SARS-CoV-2 lineages, and this revealed four additional lineages [B.1.1, B.1.36, B.1.22, B.1.1.10] of the virus circulating in Nigeria [Figure 2]. Addition of these new lineages brings the total number of lineages circulating in Nigeria to seven (7). Sequences from these lineages from Nigeria are clustering with sequences from Asia, Europe, USA, Middle-East, Australia, and other African countries [Figure 1], indicating multiple introduction of multiple lineages [Figures 1&2] of the virus into the country.
Figure 1: Maximum likelihood tree of SARS-CoV-2. Nigerian sequences are coloured green with the newly obtained sequences labelled ‘Nigeria_new’.
SARS_CoV_2_subset_09-07-20_tree.pdf (15.0 KB)
Figure 2: Lineage classification of SARS-CoV-2 in Nigeria according to genome sequence data. Lineages are grouped according to named global outbreaks of the current pandemic** (Lineage A: China, South Korea, USA; Lineage B.1: UK, USA, Australia; Lineage B.1.1: UK, Australia, USA; Lineage B.1.1.10: UK, Iceland, Australia; Lineage B.1.22: Netherlands, Australia, Austria; Lineage B.1.36: Saudi Arabia, UK, Turkey; Lineage B.2.1: UK, Australia, USA).
Genome Annotation and Mutation Analysis
We performed genome annotation on all 42 genomes from Nigeria so far using Prokka version 1.14.6 with the SARS-CoV-2 reference genome [NCBI accession number - NC_045512.2] as guide. A custom Bash script was used to extract the spike protein from each annotated output and generate a multi FASTA file containing all spike proteins. Quality control was done to exclude protein files that had missing information of up to 50 % from the analysis, yielding 26 genomes. Multiple sequence alignment using MAFFT version 7.450 shows that 19 of the 26 genomes (73.1 %) have the Glycine (G) mutation in position 614, whilst 7 genomes (26.9 %) retain the wild type Aspartic acid (D) in that position (Figure 2). This mutation has been recently been confirmed to be associated with higher transmission of SARS-CoV 2, hence dominating globally than the wild type [Korber et al. (2020)].
Figure 3: SARS-CoV-2 genomes from Nigeria showing Spike protein D614G mutation in white and orange.
Temporal Analysis of SARS-CoV-2 Spike Protein Mutation
A temporal analysis of the D614G mutation shows that G614 mutation in the spike gene has increased over time compared to D614 wild-type in Nigeria, suggesting a higher rate of transmission of mutant viruses (Figure 4).
Figure 4: Temporal trend of D614G spike protein mutation in Nigeria.
References
Katoh, K., Misawa, K., Kuma, K., & Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research , 30 (14), 3059–3066. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform | Nucleic Acids Research | Oxford Academic.
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., Hengartner, N., Giorgi, E. E., Bhattacharya, T., Foley, B., Hastie, K. M., Parker, M. D., Partridge, D. G., Evans, C. M., Freeman, T. M., de Silva, T. I., McDanal, C., Perez, L. G., Tang, H., Moon-Walker, A., … Sheffield COVID-19 Genomics Group (2020). Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell, Advance online publication. https://doi.org/10.1016/j.cell.2020.06.043.
Matranga, C. B., Gladden-Young, A., Qu, J., Winnicki, S., Nosamiefan, D., Levin, J. Z., & Sabeti, P. C. (2016). Unbiased Deep Sequencing of RNA Viruses from Clinical Samples. Journal of visualized experiments : JoVE , (113), 54117. Unbiased Deep Sequencing of RNA Viruses from Clinical Samples | Protocol.
Okonechnikov K., Golosova O., Fursov M., the UGENE team (2012). Unipro UGENE: a unified bioinformatics toolkit, Bioinformatics , 28(8), 1166–1167, Unipro UGENE: a unified bioinformatics toolkit | Bioinformatics | Oxford Academic.
Price, M. N., Dehal, P. S., & Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular biology and evolution , 26 (7), 1641–1650. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix | Molecular Biology and Evolution | Oxford Academic.
Rambaut A., Holmes E.C., Hill V., O’Toole A., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. (2020). A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv 2020.04.17.046086; doi: A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology | bioRxiv.
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England) , 30(14), 2068–2069. https://doi.org/10.1093/bioinformatics/btu.
Data availability
All sequences are available at GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria. GISAID, NCBI GenBank, and NCBI SRA accession numbers will be shared when available. We would like to thank all the authors who have kindly deposited and shared genome data on GISAID. A table with genome sequence acknowledgments can be found at GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria.
Partners and Collaborators
African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Ede, Osun State, @acegid.
Redeemer’s University, Ede, Osun State (RUN).
Nigeria Centre for Disease Control (NCDC), Abuja, Nigeria, @NCDCgov.
Irrua Specialist Teaching Hospital, Irrua, Edo State, Nigeria.
Alex Ekueme Federal University Teaching Hospital, Abakaliki, Ebonyi State, Nigeria.
Federal Medical Center-Owo, Owo, Ondo State, Nigeria.
College of Medicine, University of Ibadan, Ibadan, Nigeria.
Africa CDC, Addis Ababa, Ethiopia, @AfricaCDC.
Broad Institute and Harvard University, Cambridge, MA, USA.
Beth Israel Deaconess Medical Center, Boston, MA, USA.
Disclaimer and contact information
Please note that these analyses are based on work in progress and should be considered preliminary. Our analyses of this data are ongoing and a publication communicating our findings on these and other published genomes is in preparation. These data cannot be used without permission. If you wish to use this data please contact:
Christian Happi, PhD
Professor of Molecular Biology and Genomics, Redeemer’s University, Ede, Osun State, Nigeria
Director, African Center of Excellence for Genomics of Infectious Diseases [ACEGID]
E-mail: [email protected]
Website: www.acegid.org
Twitter: @christian_happi
Chikwe Ihekweazu, M.P.H, F.F.P.H
Director General, Nigerian Centre for Disease Control [NCDC], Abuja, Nigeria
Email: [email protected]
Website: [https://ncdc.gov.ng/]
Twitter: @Chikwe_I
Paul Eniola Oluniyi, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
E-mail: [email protected]
Twitter: @pauloluniyi
Idowu Olawoye, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
Email: [email protected]
Twitter: @idowuolawoye