Summary_MERS-CoV_15Apr15_final.pdf (573.1 KB)
Recent evolution patterns of the Middle East respiratory syndrome coronavirus (MERS-CoV)
Mazin A. Barry MD1, Sarah S. Al Subaie MD[2], Ali M. Somily MD[3], Abdulaziz A. BinSaeed, MD. PhD[4], Fahad A. Alzamil MD[5], Imad A. Al-Jahdali MD[6], Ahmed M. Alotaibi MD[7], Nahid A. Batarfi PhD[8], Scott J.N. McNabb PhD MS[9], Matthew Cotten PhD [10], Simon J. Watson PhD [10], Spela Binter MS[10], Andrew Rambaut DPhil [11,12], Paul Kellam PhD [10,13]
1 Department of Internal Medicine, Infectious Diseases, College of Medicine, King Khalid and King Saud University Hospital Riyadh, Kingdom of Saudi Arabia
2 Department of Pediatrics, Infectious Diseases Unit, College of Medicine, King Khalid and King Saud University Hospital Riyadh, Kingdom of Saudi Arabia
3 Department of Pathology, College of Medicine, King Khalid and King Saud University, Riyadh, Saudi Arabia, P.O.Box 2925, Riyadh 11461, Kingdom of Saudi Arabia
4 Department of Family and Community Medicine, College of Medicine, King Saud University, Riyadh, Saudi Arabia, P.O.Box 2925, Riyadh 11461, Kingdom of Saudi Arabia, Prince Sattam Bin Abdul Aziz Research Chair of Epidemiology and Public Health, College of Medicine, King Saud University, Riyadh Kingdom of Saudi Arabia
5 Department of Pediatrics, College of Medicine, King Khalid and King Saud University Hospital Riyadh, Kingdom of Saudi Arabia
6 Deputy Minister. Ex. General Director King Fahad General Hospital, Jeddah and Occupational and environmental medicine, Um AlQura University, Kingdom of Saudi Arabia 7 Department of Intensive Care, Prince Mohammed bin Abdulaziz Hospital, Riyadh, Kingdom of Saudi Arabia
8 Epidemiology section, Command and Control Center (CCC) Ministry of Health, Jeddah 9 Rollins School of Public Health, Emory University, Atlanta, GA, USA
10 Wellcome Trust Sanger Institute, Hinxton, United Kingdom
11 Institute of Evolutionary Biology, Ashworth Laboratories, Kings Buildings, West Mains Road, Edinburgh, United Kingdom
12 Fogarty International Center, NIH, Bethesda, Maryland, USA
13 Division of Infection and Immunity, University College London, London, United Kingdom
In a collaborative effort between the Kingdom of Saudi Arabia (KSA) Ministry of Health, King Saud University, Riyadh, King Khalid University Hospital, Riyadh, the U.S. CDC, and the Wellcome Trust Sanger Institute, we are monitoring the evolution and transmission patterns of MERS-CoV
Summary. PCR-positive upper respiratory samples from confirmed MERS cases identified from May-November 2014 in Jeddah and Riyadh, KSA were amplified and subjected to deep sequencing with paired 149 nt Illumina MiSeq data followed by de novo assembly (see (1, 2)). 22 samples yielded phylogenetically useful sequences and 8 samples yielded full MERS-CoV genomes (>28000 nt). The novel sequences are available at the following link in a simple alignment or as an alignment with all available MERS-CoV genomic data. (https://tinyurl.com/MERS-CoV-15May15)
Disclaimer. We make these sequence data publically available with the hope that they are useful for other scientists and the caveat that they are preliminary and still undergoing quality control. A manuscript analyzing the phylogenetic patterns in more detail is in preparation. If investigators would like to use these sequence data for publication prior to release of our paper, please contact us.
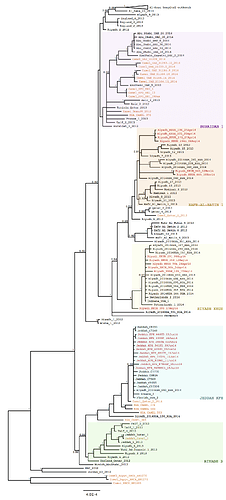
Phylogenetic patterns. The recent MERS-CoV sequences fell into three main groups (Figure 1). Six of the novel Riyadh sequences clustered in the Riyadh_KKUH clade, comprising Riyadh sequences from April 2014 (3), and Indiana_USA_2014 (obtained from a health care worker (HCW) who had worked in Riyadh and then traveled to the USA (4)). A second group of six newly generated Riyadh sequences clustered in the Hafr-al-Batin_1 (HAB_1) lineage and also included the two most recent sequences from November 2014, both from a single patient. The novel Jeddah sequences form part of the Jeddah_KFH clade and are related to Florida_USA_2014 (from a HCW who had worked in Jeddah and traveled to the USA (4)) and to other published Jeddah sequences from April 2014 (3).
Figure 1. Bayesian phylogeny of 157 MERS-CoV isolates inferred from a multiple sequence alignment of 30055 nucleotides. The phylogeny was generated using MrBayes v3.2.4, running 3 Markov chain Monte Carlo runs for 2,500,000 states under a GTR+Γ substitution model. The 22 new sequences are highlighted in red, while those isolated from camels are highlighted in orange. The Al-Hasa hospital outbreak is collapsed for clarity. Viral clades (or lineages, for HAB_1) are indicated in colored boxes. Posterior probabilities are shown for nodes with a support >0.9. The scale bar indicates the genetic distance, in substitutions/site.
The large Riyadh outbreak in the Spring 2014 appeared to be the result of multiple independent viruses (see also the blue clades 1-6 in (3)). The new sequence data identified an additional MERS-CoV source comprising Riyadh_KKUH_104_25Apr14, Riyadh_KKUH_105_25Apr14, Riyadh_KKUH_173_27Apr14, Riyadh_KKUH_294b_06May14 and possibly Riyadh_KKUH_395_17May14.
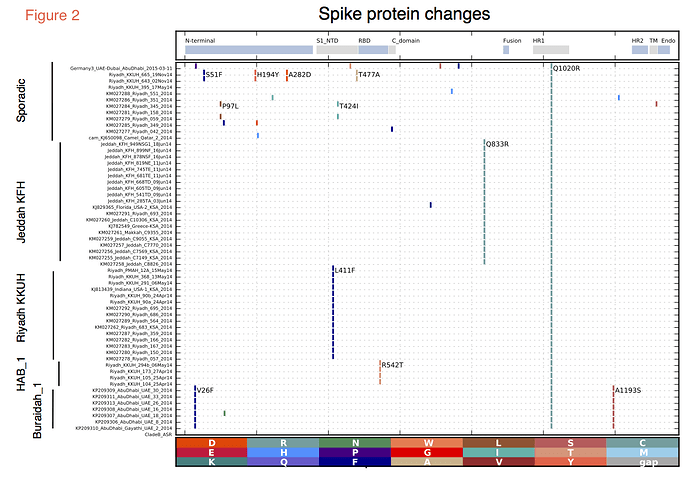
The two most recent sequences (Riyadh_KKUH_643_02Nov14 and Riyadh_KKUH_665_19Nov14) were of interest as there was a modest increase in cases reported in Riyadh in November 2014 (some with camel contact some with hospital contact). These two sequences, although clustering in the HAB_1 lineage, encode an altered spike protein with unique polymorphisms (Figure 2), suggesting a recombinant.
Spike protein changes. It is important to monitor evolution in the coronavirus spike region. Changes in the spike protein accompanied the evolution of the SARS coronavirus and may be an important marker of increased transmissibility. Multiple changes in spike protein were observed in the recent samples, with changes in or near to the receptor-binding domain (RBD) (Figure 2). An L411F change near the RBD was present in Indiana_USA_2014 and the related Riyadh sequences, while the novel Riyadh sequences in the HAB_1clade have R542T, which lies within the RBD. However, neither of these Riyadh changes appeared in the two most recent Riyadh sequences (Riyadh_KKUH_643_02Nov14, Riyadh_KKUH_665_19Nov14), which display a T477A change near the RBD. The Jeddah sequences encoded a Q833R change. None of the recent changes were found to be under positive selection using any of the selection methods within the Datamonkey suite (5).
Figure 2. Protein changes in the spike encoded by recent MERS-CoV. All recent MERS-CoV spike ORFs were translated, the proteins were aligned and amino acid differences from the reconstructed ancestral Clade B protein were determined and marked by vertical colored bars and the changes observed in more than one genome are marked within the figure. Functional domains of the spike protein are indicated in the top panel and include the N- terminal domain, the receptor binding domain, the fusion domain (Fusion), the heptad repeats 1 and 2 (HR1 and HR2), the transmembrane ™ and cytoplasmic (Endo) domain.
Evolutionary rate. The updated evolutionary rate of the virus is 8.37x10-4 (95% CI: 6.71x10-4, 1.01x10-3). From the phylogeny, the root of the tree (last common ancestor between human MERS and the Egyptian camels) is September 2010 (95% CI: November 2009, June 2011). The last common ancestor between clade A (EMC/2012 and Jordan-N3/2012) and B (rest of human isolates) is October 2010 (95% CI: January 2010, June 2011) and the root of clade B is November 2011(95% CI: June 2011, April 2012).
References
- Cotten M, Lam TT, Watson SJ, Palser AL, Petrova V, Grant P, Pybus OG, Rambaut A, Guan Y, Pillay D, Kellam P, Nastouli E. 2013. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis 19:736-742B.
- Cotten M WS, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Al Rabeeah AA, Alhakeem RF, Assiri A, Al-Tawfiq JA, Albarrak A, Barry M, Shibl A, Alrabeah FA, Hajjar S, Balkhy HH, Flemban H, Rambaut A, Kellam P, Memish ZA. 2014. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5:e01062-01013.
- Drosten C, Muth D, Corman VM, Hussain R, Al Masri M, HajOmar W, Landt O, Assiri A, Eckerle I, Al Shangiti A, Al-Tawfiq JA, Albarrak A, Zumla A, Rambaut A, Memish ZA. 2015. An observational, laboratory-based study of outbreaks of middle East respiratory syndrome coronavirus in Jeddah and Riyadh, kingdom of Saudi Arabia, 2014. Clin Infect Dis 60:369-377.
- Bialek SR, Allen D, Alvarado-Ramy F, Arthur R, Balajee A, Bell D, Best S, Blackmore C, Breakwell L, Cannons A, Brown C, Cetron M, Chea N, Chommanard C, Cohen N, Conover C, Crespo A, Creviston J, Curns AT, Dahl R, Dearth S, DeMaria A, Echols F, Erdman DD, Feikin D, Frias M, Gerber SI, Gulati R, Hale C, Haynes LM, Heberlein-Larson L, Holton K, Ijaz K, Kapoor M, Kohl K, Kuhar DT, Kumar AM, Kundich M, Lippold S, Liu L, Lovchik JC, Madoff L, Martell S, Matthews S, Moore J, Murray LR, Onofrey S, Pallansch MA, Pesik N, Pham H, et al. 2014. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities - May 2014. MMWR Morb Mortal Wkly Rep 63:431-436.
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455- 2457.