Hi,
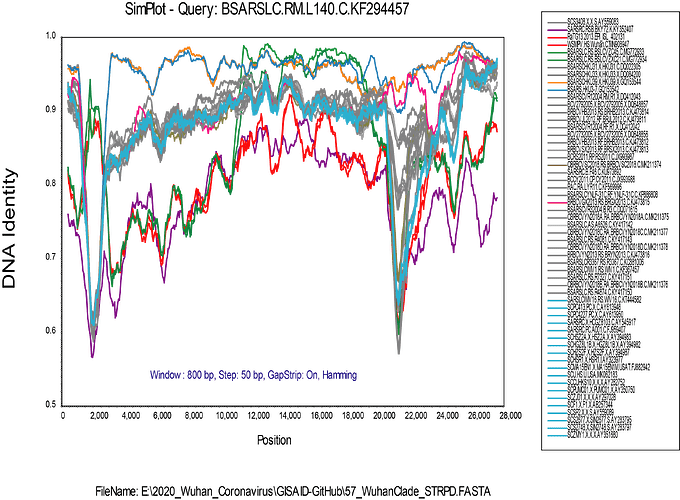
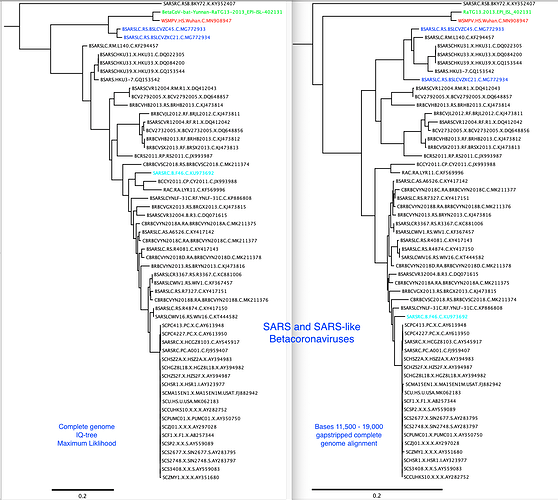
I have added the 2013 Yunnan bat sequence (much more closely related to the Wuhan2019-2020 virus than are the XC21 and ZC45 viruses) and built trees from complete genomes and some subgenomic regions chosen based on similarity plots.
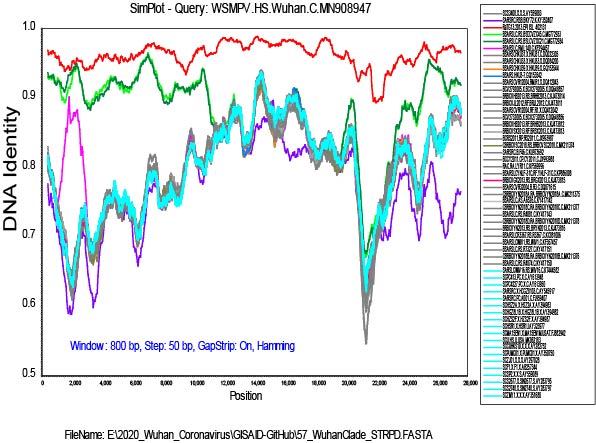
From this type of data, we cannot say which viruses are “recombinant” and which are “pure”
or never recombined in their history. It is only clear that recombination has been happening in the evolution of these viruses.
My alignment is built from a sampling of complete genomes of only the SARS and “SARS-like” subclade of the Betacoronaviruses. It does not include MERS or other Betacoronaviruses. For the SimPlot (Stuart Ray software) I first gap-stripped the alignment.
I am now running an RDP4 (Darren Martin software) alignment on the gap-stripped set, and it is finding a lot of recombination. The recombination events that I found evident in SimPlot are also detected in RDP4, but I am a new user of RDP4 and it will take me a while to get up to speed with using it.
Brian Foley
HIV Databases at LANL