MPXV intrahost variation in the context of APOBEC deamination: An initial look
Anton Nekrutenko | galaxyproject.org, Penn State, CEFE CNRS
APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) converts C in single stranded stretches of DNA or RNA to U resulting C → T transition. Members of APOBEC family target specific sequence motifs: TC , CCC , TTC where the last base mutated to T , or TCW ( W = A / T ) where the middle base mutates to T (see Chen and MacCarthy 2014 ).
Recently O’Toole & Rambaut conducted a survey of potential APOBEC signatures in a set of MPXV genomes. If APOBEC is indeed partially responsible for introducing nucleotide changes to MPXV genome in humans, we should be able to observe these changes in intrahost samples with intermediate alternative allele frequencies.
Data
The challenge is, of course, finding these data. Despite the fact that a number of datasets have been deposited to EBI SRA, recent indexing issues at EBI prevent download of these data. At the time of writing (June 13, 2022) the only available (the ones that can actually be downloaded) SRA datasets were:
| BioProject | Description | Number of samples | Platform |
|---|---|---|---|
| PRJNA844567 | Second draft genome from Spain of the Monkeypox virus 2022 outbreak | 1 | ONT |
| PRJNA842892 | Monkeypox virus from ongoing epidemic in UK (May-2022) | 4 | ONT |
| PRJNA844330 | Full-Length Genome characterization of a monkeypox case in Northeast Italy | 1 | Illumina |
| PRJNA845087 | Illumina whole-genome sequence of Monkeypox virus in a patient traveling from the Canary Islands to France | 4 | Illumina |
Since Illumina data is more suitable for detection of low frequency variants (5% and up) we analyzed data provided by PRJNA845087 (also described here).
Analysis
We analyzed PRJNA845087 using Galaxy workflows that have also been used in this study. The analysis artifacts can be examined and/or downloaded here:
| Artifact | Description | Link |
|---|---|---|
| Galaxy History | A complete record of the analysis with all starting and intermediate datasets | Galaxy |
| Variant list | A tab-delimited list of variants used in this analysis | [dataset] |
Results
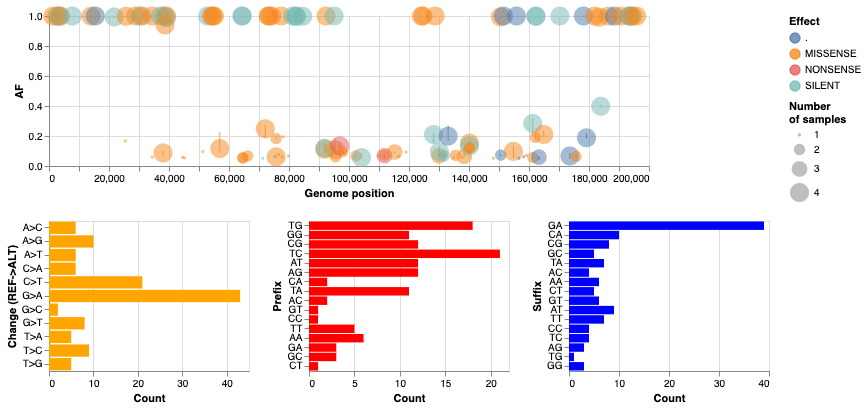
The following image provides quick summary of the results. The interactive version of this image is here.
- The majority of changes are
C->TandG->A - While most of these changes are fixed, some exist in intermediate frequencies (can be seen in the interactive version).
- Some change classes (e.g.,
T->C) only exist in intermediate frequencies
We are waiting on more datasets becoming available to expand the interpretation of these results.