First mpox virus clade Ib cases in Goma, Nord Kivu, DRC: Potential risk for a larger outbreak
Since the identification of the first human case of mpox in 1970, the Democratic Republic of the Congo (DRC) has borne the greatest burden of disease among endemic countries, which experiencing sporadic outbreaks almost every year. From January 2023, the DRC has seen a drastic increase in the number of suspected mpox cases, with an estimated 20,000 cases and 1,000 deaths reported at the time of writing, marking a more than 300% increase compared to previous years [1]. The disease remains concentrated in several geographic areas, primarily infecting children under 15 years of age. In recent months an explosion of cases have been detected in DRC’s regions previously not affected, coinciding with reported transmission through sexual contact [2-4].
The province most affected by these shifting transmission patterns is Sud Kivu. Since October 2023, the province has experienced a major outbreak originating from Kamituga health zone, a mining area with a substantial sex industry. This outbreak is caused by a new variant of Clade I mpox virus (MPXV). Genomic analysis of the circulating strain revealed multiple mutations induced by the human apolipoprotein B mRNA editing enzyme, catalytic subunit (APOBEC3) cytosine deamination activity, a hallmark sign of sustained human-to-human transmission. Based on distinct genetic differences with other circulating strains in the DRC, it was proposed to denote this variant as subclade Ib [2].
Since October 2023, case numbers have been increasing exponentially, and the outbreak has expanded further from Kamituga to other health zones in Sud Kivu [2, 4]. At the start of the epidemic, 10 suspected cases were notified per week. In epidemiological week 24 of 2024, there have been more than 160 mpox-suspected cases reported per week in Sud Kivu [5].
The presence of mpox in Sud Kivu with sustained human-to-human transmission poses a significant risk of a large epidemic that could spread to neighboring provinces and other countries in the Great Lakes sub-region, such as Tanzania, Rwanda, Burundi and Uganda.
In early June, an individual living in the Nyiragongo health zone of Goma, Nord Kivu, presented with skin lesions and was confirmed Mpox positive (Goma patient 1). The individual reported having recently stayed in Bukavu, in Sud Kivu, which is currently affected by the ongoing clade Ib MPXV outbreak. He also reported first symptoms on May 24th, 2024. A sample was collected on the June 1st, 2024 , and tested positive for orthopoxvirus using GeneXpert PCR platform at the Rodolphe Mérieux INRB Goma Laboratory. A second patient (Goma patient 2) from Nyiragongo without established epidemiological link with the first patient (Goma patient 1) developed symptoms on June 8th, 2024. He consulted at a health center and was referred to hospital for dermatological treatment. Samples were collected on June 14th, 2024 and tested Mpox positive.
On June 15th, 2024, a third person (Goma patient 3) was investigated. The patient presented mpox suspect lesions and reported symptoms onset June 5th, 2024. The individual resided in Goma Health zone and came from Bulengo, a displaced person site, and had no apparent epidemiological link to the previous cases.
Details about sequencing
Crust lesions, nasopharyngeal secretions and blood samples collected from the patients. PCR testing was performed using the GeneXpert with Clade II and Orthopox cartridges according to the manufacturer instructions, and a sample with amplification Ct value less than 39 was concluded positive, according to preliminary WHO guidance.
For sequencing, 3 first positive samples were extracted and sequenced. Multiplex PCR amplifications were performed using MPXV clade IIb-specific primers (PrimalScheme for reference genome MT903345), which generate 2.5 kb amplicons covering the complete MPXV genome. The PCR products were then quantified using Qubit device with QubitTM 1X dsDNA High Sensitivity Assay kit (ThermoFisher Scientific). The sequencing libraries were prepared using the rapid barcoding kit (SQK-RBK110.96; Oxford Nanopore Technologies (ONT), UK), following the manufacturer’s instructions. Sequencing libraries were loaded on R9.4.1 flow cells and run on a GridION.
Twelve additional individuals were confirmed mpox positive in the city of Goma. Notably, eight cases were in the displaced persons camp in Mudja, posing a significant risk of disease spreading this vulnerable population, in the city of Goma, accross the country and even in neighboring countries. Here, we describe the results of the genomic analysis of the strains isolated from these three initial cases.
RESULTS
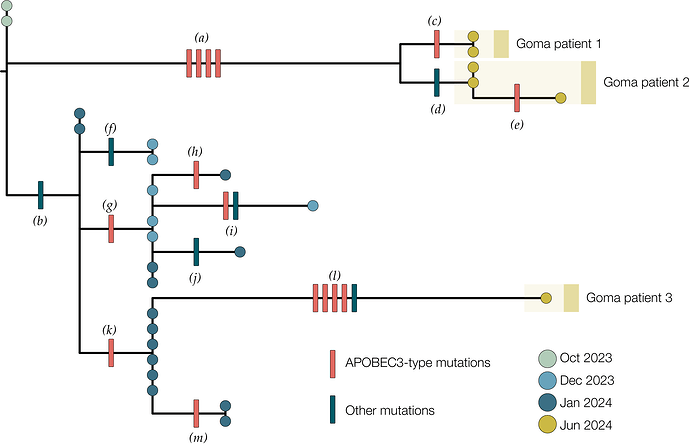
Genomic analysis of the first three confirmed cases showed that they all clustered within MPXV Clade Ib together with Mpox cases detected in Sud Kivu (Figure 1). Their position in the Sud Kivu tree suggests they were part of the sustained human outbreak first reported in Kamituga health zone. These findings are consistent with the reported travel of the first case from Sud Kivu. The genomes from the first and second cases group together suggesting they are part of the same transmission chain. The third sequenced case is separated from the first two in the Clade Ib outbreak tree, implying an independent introduction into Goma.
Figure 1 | A maximum likelihood tree constructed using IQ-Tree 2 [6] with the HKY substitution model [7]. We included a Clade Ia genome as an outgroup and then removed it after rooting. Single nucleotide mutations are reconstructed and displayed denoting whether they are APOBEC3-mediated (red bars) or other mutations (blue bars). Genomes from recent cases in Goma, Nord Kivu are denoted with yellow circles. Patients 1 and 2 each have multiple genomes sequenced from different samples. Blue circles are genomes from Kamituga, Sud Kivu from October and December 2023, and January 2024. Letters in parentheses are referenced in Table 1.
Of the 21 single nucleotide mutations reconstructed in this tree (Table 1), 15 are of the type expected due to the action of human APOBEC3 [8]. The viruses sequenced from patients 1 & 2 are descendants of viruses present in Kamituga in October 2023 or earlier. Furthermore, the branch leading to patient 1 & 2 has four APOBEC3-type mutations which even at the elevated rate of evolution induced by APOBEC3 would represent months of human transmission. In combination with the report of recent travel history means this is likely a recent introduction into the Goma area from an area of Sud Kivu with ongoing transmission.
The branch leading to patient 3 has four APOBEC3 mutations and one other mutation, possibly the result of an error during replication, indicating a similar timespan for this branch. However, this branch joins the Kamituga tree clustering with viruses sampled in January 2024, suggesting a direct link to that outbreak. The long unbroken branch with 5 single nucleotide mutations, and the fact that this lineage has not yet been sampled in Kamituga, may mean that the virus has been circulating in Goma for weeks or months.
Table 1 | List of mutations in the 2023-2024 DRC mpox outbreak.
| Branch label1 | Genome position2 | Mutation3 | APOBEC3? |
|---|---|---|---|
| (a) | 103,660 | GA->AA |
yes |
| 130,155 | GA->AA |
yes | |
| 142,587 | GA->AA |
yes | |
| 161,978 | TC->TT |
yes | |
| (b) | 165,597 | C->A |
no |
| (c) | 64,117 | GA->AA |
yes |
| (d) | 30,660 | T->C |
no |
| (e) | 181,268 | GA->AA |
yes |
| (f) | 167,805 | A->G |
no |
| (g) | 115,484 | GA->AA |
yes |
| (h) | 57,332 | GA->AA |
yes |
| (i) | 89,000 | GA->AA |
yes |
| 127,619 | A->G |
no | |
| (k) | 183,181 | G->A |
no |
| (l) | 71,751 | TC->TT |
yes |
| 139,485 | TC->TT |
yes | |
| 148,221 | GA->AA |
yes | |
| 168,749 | GA->AA |
yes | |
| 169,501 | T->A |
no |
1 Branch labels refer to Figure 1
2 Coordinates relative to Clade I reference genome, NCBI accession NC_003310
3 Dinucleotide mutations are ascribed to APOBEC3 mutations
CONCLUSION
The detection of mpox cases in the Nord Kivu highlights the importance of reinforcing surveillance measures additionally, the role of contact tracing and public health measures in controlling the spread of emerging infectious diseases in neighboring provinces and countries. Therefore, there is an urgent need for collaborative efforts and actions to combat mpox before its further spread to other countries.
DATA AVAILABILITY
The genome sequences are available on demand. Please contact Dr Eddy Kinganda, Prof Steve Ahuka or Prof Placide Mbala for coordination.
PARTNERS AND COLLABORATORS
The work is accomplished through the continuous, joined and coordinated efforts of partners supporting INRB and Mpox surveillance activities at the National level. Below are involved institutions :
- Institut National de Recherche Biomédicale (INRB), Kinshasa, Democratic Republic of the Congo : Adrienne Amuri-Aziza, Eddy Kinganda-Lusamaki, Emmanuel Hasivirwe Vakaniaki, Emmanuel Lokilo, Elisabeth Pukuta-Simbu, Jean-Claude Makangara-Cigolo, Gradi Luakanda, Olivier Tshiani, Prince Akil-Bandali, Sifa Kavira, Tony Wawina-Bokalanga, Steve Ahuka-Mundeke, Jean-Jacques Muyembe-Tamfum, Placide Mbala-Kingebeni
- Rodolphe Merieux INRB-Goma Laboratory, Goma, Democratic Republic of the Congo : Brigitte Modadra-Madakpa, Noella Mulopo-Mukanya, Michel Ngimba, Zephanie Paluku Kalimuli, Adèle Kavira Kamaliro, Emile Muhindo-Milonde, Jeriel Mufungizi, Hugo Kavunga, Tavia Bodisa-Matamu, Yves Birindwa Hamisi, Daniel Mukadi-Bamuleka
- Institute of Ecology and Evolution, University of Edinburgh, Edinburgh EH9 3FL, UK: Áine O’Toole, Andrew Rambaut
- Department of Clinical Sciences, Institute of Tropical Medicine, Antwerp, Belgium: Daan Jansen, Laurens Liesenborghs, Tony Wawina-Bokalanga, Koen Vercauteren
- Department of Epidemiology, Infectious Diseases and Microbiology, University of Pittsburgh, School of Public Health, Pittsburgh, PA, USA: Jean Nachega
- Hemorrhagic Fever and Monkeypox Program, Ministry of Health, Kinshasa, Democratic Republic of the Congo : Cris Kacita, Robert Shongo
- TransVIHMI (Recherches Translationnelles sur le VIH et les Maladies Infectieuses), Université de Montpellier, Institut de Recherche pour le Développement (IRD), Institut National de Santé et de Recherche Médicale (INSERM), Montpellier, France : Ahidjo Ayouba, Eddy Kinganda-Lusamaki, Martine Peeters, Eric Delaporte
- Provincial Health Division, North Kivu, Democratic Republic of the Congo : Gaston Lubambo Maboko, Guy Mutombo Ndongala
- WHO : Alain Kakule Mangolopa, Amos Kiuka Ebondo, Lorenzo Subissi
- Department of Epidemiology, Jonathan and Karin Fielding School of Public Health, University of California, Los Angeles, CA, USA : Nicole A. Hoff, Sydney Merritt, Anne Rimoin
- Service de Microbiologie, Département de Biologie Médicale, Cliniques Universitaires de Kinshasa, Université de Kinshasa : Raphael Lumembe, Gabriel Kabamba, Placide Mbala-Kingebeni, Tony Wawina-Bokalanga, Jean-Jacques Muyembe-Tamfum, Steve Ahuka-Mundeke
- US Department of Agriculture, Manhattan, KS, USA : Lisa E. Hensley
- University of Manitoba, Winnipeg, Manitoba, Canada : Jason Kindrachuk
- University of Bern : Nicola Low
- University of Birmingham, Birmingham, UK : Sam Wilkinson and Nicholas Loman
- Africa CDC : Sofonias Kifle Tessema
STATEMENT ON CONTINUING WORK AND ANALYSES PRIOR TO PUBLICATION
These genome sequences are being shared pre-publication. Please note that this data is based on work in progress and should be considered preliminary. Our analyses of these data are ongoing and a publication communicating our findings on these, and other published genomes is in preparation. If you intend to use these sequences prior to our publication, please communicate with Dr Eddy Kinganda, Prof Steve Ahuka and Prof Placide Mbala for coordination.
COMPETING INTERESTS
None of the other authors declare competing interests.
FUNDING
This work was funded by the Department of Defense, Defense Threat Reduction Agency, Monkeypox Threat Reduction Network; and USDA Non-Assistance Cooperative Agreement #20230048; Africa Pathogen Genomics Initiative helped acquiring and maintaining the sequencer; Agence Française de Dévelopement through the AFROSCREEN project (grant agreement CZZ3209, coordinated by ANRS-MIE Maladies infectieuses émergentes in partnership with Institut de Recherche pour le Développement (IRD) and Pasteur Institute) for laboratory support and PANAFPOX project funded by ANRS-MIE ; Belgian Directorate-general Development Cooperation and Humanitarian Aid and the Research Foundation - Flanders (FWO, grant number G096222 N to L.L.); the International Mpox Research Consortium (IMReC) through funding from the Canadian Institutes of Health Research and International Development Research Centre (grant no. MRR-184813); US NIAID/NIH grant number U01AI151799 through Center for Research in Emerging Infectious Disease-East and Central Africa (CREID-ECA); E.L. received a PhD grant from the French Foreign Office. We acknowledge the support of the Wellcome Trust (Collaborators Award 206298/Z/17/Z, ARTIC network)
REFERENCES:
-
Mbala-Kingebeni, P., et al. (2024) The time is now (again) for mpox containment and elimination in Democratic Republic of the Congo. PLOS Glob Public Health 4: e0003171.
-
Vakaniaki, E.H., et al. (2024) Sustained Human Outbreak of a New MPXV Clade I Lineage in the Eastern Democratic Republic of the Congo. Nat Med, in press.
-
Kibungu, E.M., et al. (2024) Clade I-Associated Mpox Cases Associated with Sexual Contact, the Democratic Republic of the Congo. Emerg Infect Dis 30: 172-176.
-
Masirika, L.M., et al. (2024) Ongoing mpox outbreak in Kamituga, South Kivu province, associated with monkeypox virus of a novel Clade I sub-lineage, Democratic Republic of the Congo. Euro Surveill 29.
-
INSP-RDC (2024) Rapport de situation No. 22, Données de la semaine épidémiologique 24 Période du 17-23 Juin 2024
-
Minh, B.Q., et al. (2020) IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era.Mol Biol Evol 37: 1530-1534.
-
Hasegawa, M., et al. (1985) Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160-174.
-
O’Toole, A., et al. (2023) APOBEC3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016. Science 382: 595-600.