The African Centre of Excellence for the Genomics of Infectious Disease (ACEGID) at Redeemer’s University (RUN), Ede, Nigeria, reports the first genome sequences of Rabbit Haemorrhagic Disease virus (RHDV) in Sub-Saharan Africa. These sequences are available at: GitHub - acegid/RHDV_sequences: Rabbit hemorrhagic disease virus (RHDV) sequences from Nigeria.
In August 2020, following reports of devastating outbreaks of suspected Rabbit Haemorrhagic Disease (RHD) in Rabbit farms around, Ibadan, South-western region of Nigeria, tissue samples were obtained from five rabbits post mortem for molecular confirmation. Symptoms observed included: anorexia, lacrimal discharge, lethargy, bleeding from the oral and nasal orifices and sudden death in mostly exotic breeds of all ages and sex.
On the 25th September, 2020, at ACEGID, samples were homogenized in TRIzol and nucleic acid extracted using the QIAamp Viral RNA extraction kit (Qiagen, Hilden, Germany). RT-PCR targeting the VP60 gene of the RHDV genome was carried out using One-step SuperScript™ III One-Step RT-PCR with Platinum™ Taq DNA Polymerase (Invitrogen, USA) kit. RT-PCR products were viewed in 1% agarose gel. Two out of five samples were RT-PCR positive for RHDV. Nextera XT sequencing libraries were prepared from extracted RNA following previously published protocol (Matranga et al., 2016).
Libraries were sequenced using the Illumina Miseq machine in the sequencing platform of ACEGID. Following sequencing, raw reads from the next-generation sequencing machine were uploaded to our cloud-based platform (DNAnexus, www.dnanexus.com). Quality control check was conducted on the raw reads using fastqc (Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data). Metagenomics analysis was carried out using Kraken2 (Wood et al., 2019). Three RHDV genomes (One full and two partial) were assembled on the 1st October, 2020 using our publicly-available software viral-ngs v2.1.8 (GitHub - broadinstitute/viral-ngs: Viral genomics analysis pipelines) implemented on DNAnexus. Sequence analysis revealed that the three sequences from this study belong to genotype Lagovirus europaeus/GI.2 also known as RDHV2 or RHDVb. This genotype was first identified in 2010 as a novel pathogenic form of lagovirus in France (Le Gall-Reculé et al., 2011) after which it spread rapidly through Europe and other parts of the world (Oceania, North America and Africa). It has also been reported to replace former circulating GI.1 strain in Australia and Portugal (Lopes et al., 2015; Mahar et al., 2018).
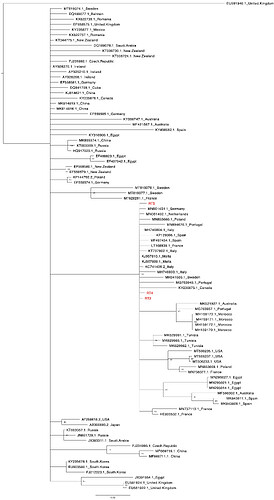
Ninety representative RHDV sequences were obtained from the NCBI database and aligned with our three sequences using MAFFT v7.388 (Katoh et al., 2002) with further adjustment made manually as necessary in Geneious (Kearse et al., 2012). A neighbour-joining tree was generated also in Geneious from a distance matrix corrected for nucleotide substitutions by the Tamura-Nei model. The tree was viewed and manually edited using FigTree (FigTree). Phylogenetic analysis showed that the sequences from this study belong to the RHDV2 genotype as they clustered together in the same clade with previous RHDV2 sequences from Europe (Figure 1).
To investigate the discrepancy between the PCR and sequencing results, we aligned the three sequences from this study with all sequences in the same RHDV2 clade on the tree to check for amino acid mutations specific to our new sequences from Nigeria. We also mapped the RT-PCR primers to our full genome obtained from this study to check if any mutation observed occurred in the target regions of the primers. Analysis of the Single Nucleotide Polymorphisms (SNPs) in the RHDV2 genome of a sample (RT5) obtained from our study, revealed 11 unique mutations resulting in amino acid changes. These are: Thr179Ile, Leu216Ser, Leu342Ser, Arg827Lys, Gln928Arg, Thr1337Ile, Thr1742Ile, Leu1929Pro, Ser1978Phe, Val2104Ala, Val2127Ala. Four of these mutations (Leu1929Pro, Ser1978Phe, Val2104Ala, Val2127Ala) occur in the region of target for the RT-PCR primers. Sample RT4 had a unique mutation Pro440Leu compared to other RHDV2 sequences they clustered with on the tree.
Figure 1: Phylogenetic relationship between the sequences from this study (coloured red)
and representative RHDV sequences obtained from the NCBI database
Clearer version of the phylogenetic tree is available at this link (RHDVtree.pdf (4.8 KB))
References
Matranga, C. B., Gladden-Young, A., Qu, J., Winnicki, S., Nosamiefan, D., Levin, J. Z., & Sabeti, P. C. (2016). Unbiased Deep Sequencing of RNA Viruses from Clinical Samples. Journal of visualized experiments : JoVE , (113), 54117. Unbiased Deep Sequencing of RNA Viruses from Clinical Samples (Video) | JoVE | Protocol.
Wood, D.E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2.Genome Biol 20, 257 (2019). Improved metagenomic analysis with Kraken 2 | Genome Biology | Full Text.
Le Gall-Reculé G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guérin JL, Marchandeau S. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet Rec. 2011 Feb 5;168(5):137-8. doi: 10.1136/vr.d697. PMID: 21493491.
Lopes, A.M.; Correia, J.; Abrantes, J.; Melo, P.; Ramada, M.; Magalhães, M.J.; Alves, P.C.; Esteves, P.J. Is the New Variant RHDV Replacing Genogroup 1 in Portuguese Wild Rabbit Populations? Viruses 2015, 7, 27-36.
Mahar JE, Hall RN, Peacock D, Kovaliski J, Piper M, Mourant R, Huang N, Campbell S, Gu X, Read A, Urakova N, Cox T, Holmes EC, Strive T. 2018. Rabbit hemorrhagic disease virus 2 (RHDV2; GI.2) is replacing endemic strains of RHDV in the Australian landscape within 18 months of its arrival. J Virol 92:e01374-17. https://doi.org/10.1128/JVI.01374-17.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002 Jul 15;30(14):3059-66. doi: 10.1093/nar/gkf436. PMID: 12136088; PMCID: PMC135756.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012 Jun 15;28(12):1647-9. doi: 10.1093/bioinformatics/bts199. Epub 2012 Apr 27. PMID: 22543367; PMCID: PMC3371832.
Data availability
All sequences are available at GitHub - acegid/RHDV_sequences: Rabbit hemorrhagic disease virus (RHDV) sequences from Nigeria. NCBI GenBank accession numbers(MW123059 - MW123061) were received on 16th, October, 2020.
Partners and Collaborators
African Centre of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Ede, Osun State, @acegid_lab
Redeemer’s University, Ede, Osun State, Nigeria
Department of Veterinary Pathology, University of Ibadan, Ibadan, Nigeria
Laboratory of Viral Zoonotics, Department of Veterinary Medicine, University of Cambridge, Cambridge, UK.
Disclaimer and contact information
Please note that these analyses are based on work in progress and should be considered preliminary. Our analyses of this data are ongoing and a publication communicating our findings on these and other published genomes is in preparation. If you wish to use this data please contact:
Christian Happi, PhD
Professor of Molecular Biology and Genomics, Redeemer’s University, Ede, Osun State, Nigeria
Director, African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
E-mail: [email protected]
Website: www.acegid.org
Twitter: @christian_happi
Anise Happi, DVM, MVSc, PhD
Department of Veterinary Pathology
Faculty of Veterinary Medicine
University of Ibadan, Ibadan, Nigeria.
Email: [email protected]
Website: www.acegid.org
Jonathan L. Heeney, DVM, PhD
Laboratory of Viral Zoonotics
Department of Veterinary Medicine,
University of Cambridge, Cambridge, UK.
Email: [email protected]
Website: www.vet.cam.ac.uk/
Twiter: @JonathanHeeney
Olusola A. Ogunsanya, DVM, (MVSc in view)
Department of Veterinary Pathology,
Faculty of Veterinary Medicine,
University of Ibadan, Ibadan, Nigeria.
Email: [email protected]
Website: www.acegid.org
Judith Uche Oguzie, DVM, MSc, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
Email: [email protected]
Website: www.acegid.org
Twitter: @judith_oguzie
Paul Eniola Oluniyi, MSc, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
E-mail: [email protected]
Website: www.acegid.org
Twitter: @pauloluniyi