Continuing the discussion from Latest tree 17th Feb (2 more Brazilian genomes):
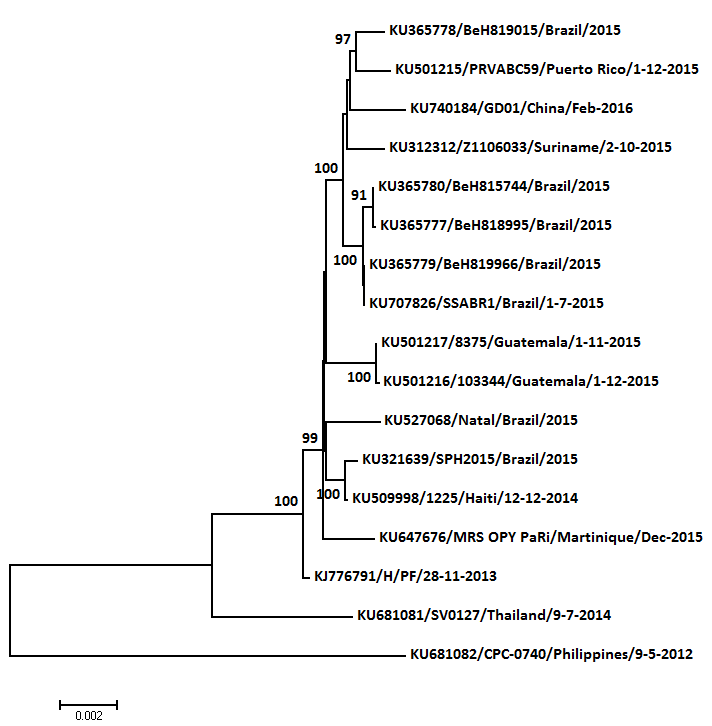
One more genome this week, a traveller returning to China (from Brazil by the look of it).
NJ, T93+G, midpoint rooted, 2 randomly chosen SE Asian genomes also included to avoid forcing root on Polynesia
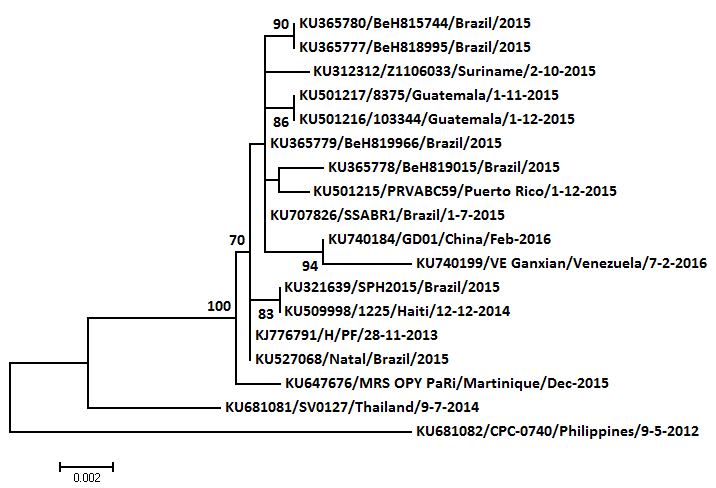
Other Chinese sequence, this one a fragment, published and of Venezuelan origin, is nearest neighbour of (but not identical to) unpublished full-length Chinese genome. K2, ML (only a couple of kb in alignment), mid-point root, SE Asians included to avoid root-force
Dear Derek - would it be possible for you to share the latest alignment for this tree for the purposes of developing a multiplex amplicon scheme for Zika sequencing?
Many thanks!
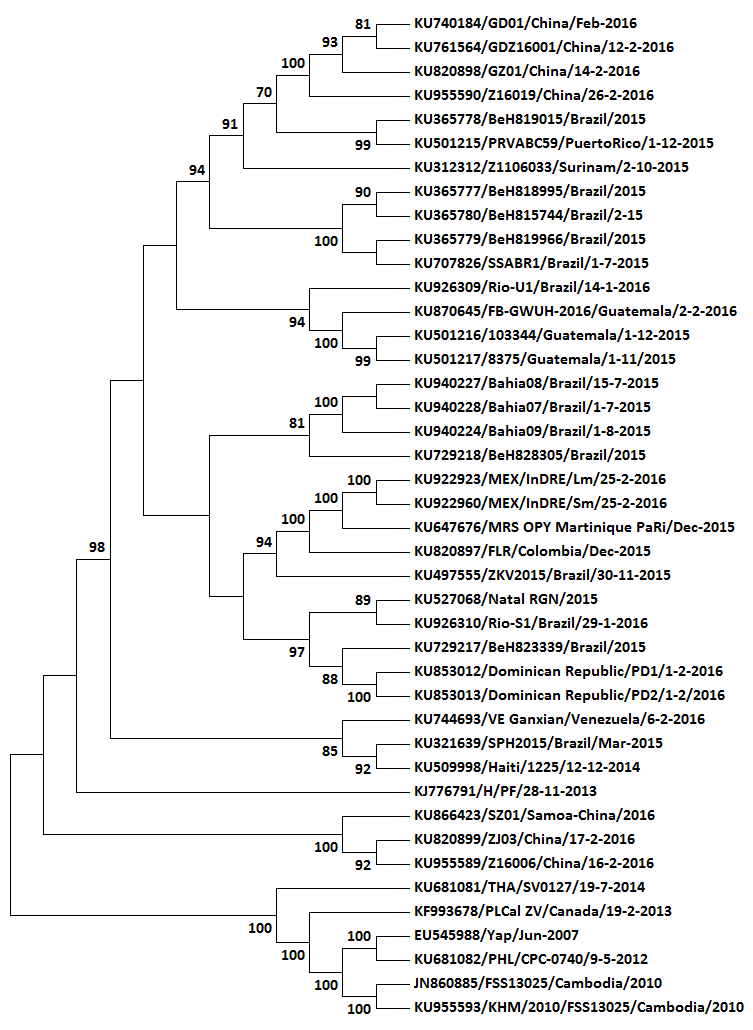
This is the tree of the latest alignment. I’ll try to get the actual alignment to you soon (Virological won’t take a *.mas attachment, so some rendering to *.txt will be required)
Hope this is downloadable, and usable once downloaded…
Zika_APA_noMalays_Mar_2016.fasta.txt (445.5 KB)