Overview
https://nextstrain.org/ is a widely used application for visualizing pathogen evolution. It has been a critical resource during the analysis of SARS-CoV-2 community spread. The Nexstrain dashboard is powered by Auspice, the open-source library used for visualization.
Recently, on the JBrowse 2 team, we have released a package on NPM that enables embedding the JB2 linear genome browser into a ReactJS application. In order to provide a resource for SARS-CoV-2 analysis, we have embedded the genome browser into an instance of Auspice loaded with SARS-CoV-2 data.
Demonstration
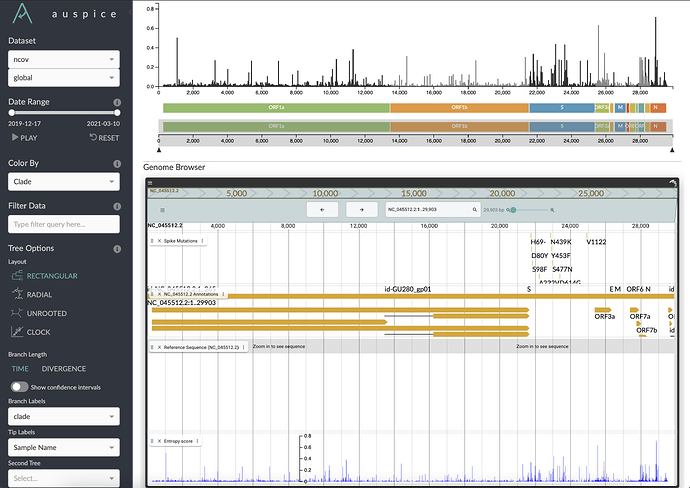
To augment the existing Diversity Panel in Nexstrain, we have embedded a JB2 genome browser in Auspice. We have loaded the browser with additional annotation data, such as the NCBI RefSeq annotations, and a track annotating mutations to the Spike glycoprotein:
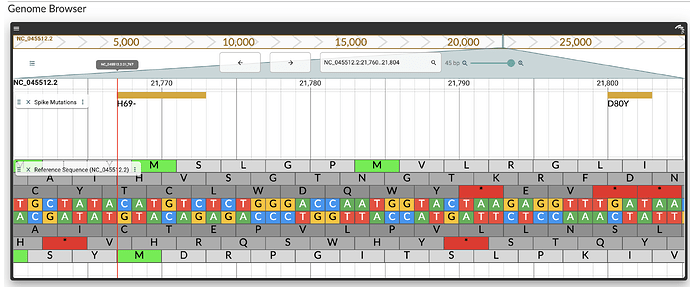
It is also possible to zoom and pan in the browser. When zoomed in at a sufficient resolution, the SARS-CoV-2 reference sequence is also displayed, with translation frames:
Resources
The URL for the tool is: https://nextstrain-jbrowse.herokuapp.com/ncov/global
The source code is available at: GitHub - GMOD/auspice: Web app for visualizing pathogen evolution
Additionally, we have added example code and data to our package documentation for anybody else that wants to embed a SARS-CoV-2 genome browser in their own application:
Interactive documentation for the JB2 React component including the Nextstrain example will be available online in the next JBrowse 2 release.