[Update information]
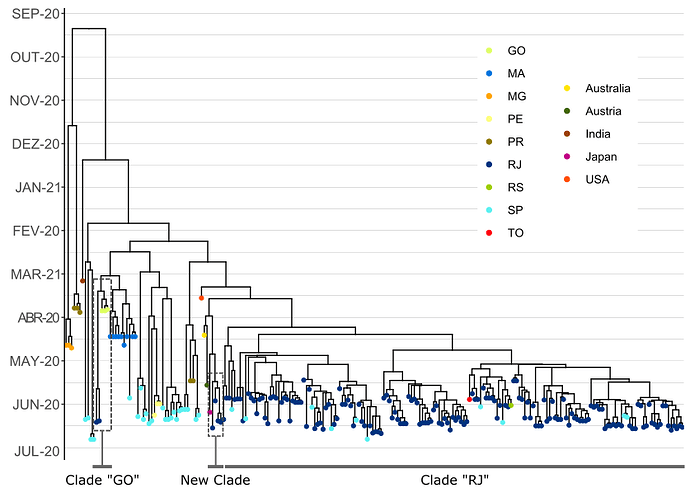
We have sequenced 96 additional samples of B.1.617.2 lineage samples collected in Rio de Janeiro between June, 22th, 2021 and July, 19th 2021 as part of the Corona-ômica-RJ Network surveillance project (see the update on https://pando.tools/t/genomic-surveillance-of-sars-cov-2-in-the-state-of-rio-de-janeiro-brazil-technical-briefing/683/6). Evolutionary analysis of these sequences has grouped them within three clades. The first clade is composed of the B.1.617.2 genomes from Rio de Janeiro previously sequenced and described in this post. Besides the already identified dispersal of this clade to São Paulo, B.1.617.2 sequences from the states of Tocantins and Rio Grande do Sul also are located within it. A second clade is composed by a new introduction in the country, with the first divergence between Brazilian sequences estimated to have happened mid-May. The third clade is formed by sequences related to genomes sampled in the Brazilian state of Goiás. The dispersal of this last clade within Rio de Janeiro is estimated to have also happened in mid-May.
**
Figure 1. Time tree of Brazilian genomes from the B.1.617.2 (Delta) lineage. Global sequences were added in order to test the evolutionary relationship of the new introduction to the clade previously identified in the state of Rio de Janeiro (RJ). Colored dots represent the sampling location of the sequences and abbreviations correspond to Brazilian states: Goiás (GO), Maranhão (MA), Minas Gerais (MG), Pernambuco (PE), Paraná (PR), Rio Grande do Sul (RS), São Paulo (SP) and Tocantins (TO).
We would like to thank all the authors and administrators of the GISAID database, which allowed this study of genomic epidemiology to be conducted properly. A full list acknowledging the authors publishing the data used in this update can be found in the following file:
Acknowledgement_GISAID.pdf (37.0 KB)