First monkeypox virus genome sequence from Brazil
Ingra Morales Claro, Evelyn Lepka de Lima, Camila Malta Romano, Darlan da Silva Candido, José Angelo Lauletta Lindoso, Luiz Alberto Costa Barra, Luciana Marques Sansão Borges, Lucas Alberto Medeiros, Marcia Y. S. Tomishige, Mariana Severo Ramundo, Tomas Moutinho, Anderson José Dias da Silva, Camila Cristina Martini Rodrigues, Luiz Cesar Fernandes de Azevedo, Lucy Santos Villas-Boas, Camila A. M. da Silva, Thaís M. Coletti, Áine O’Toole, Joshua Quick, Nick Loman, Andrew Rambaut, Nuno R. Faria, Claudia Figueiredo-Mello, Ester C. Sabino
Note: This work now been published as I. M. Claro, C. M. Romano, D. D. S. Candido, E. L. Lima, J. A. L. Lindoso, M. S. Ramundo, et al. Shotgun metagenomic sequencing of the first case of monkeypox virus in Brazil, 2022. Rev Inst Med Trop Sao Paulo 2022 Vol. 64 Pages e48. https://www.ncbi.nlm.nih.gov/pubmed/35749419 DOI: 10.1590/S1678-9946202264048.
Monkeypox virus (MPXV) is a double-stranded DNA virus with a 197-kb genome. MPXV is a member of the Orthopoxvirus (OPV) genus from the Poxviridae family, which also includes variola virus that causes smallpox (ICTV, 2011). Monkeypox was first identified in humans in 1970 in the Democratic Republic of Congo. Since then, several outbreaks of monkeypox have been reported in the African continent (Bunge et al. 2022), where 1408 suspected cases and 66 deaths were reported since the beginning of 2022 (World Health Organization 2022). MPXV is currently classified by phylogenetic analysis into two lineages, the West and Central African clade, although a novel classification of MPXV into MPXV Clades 1, 2 and 3 has recently been proposed (Happi et al., 2022). Newly proposed MPXV Clade 3 includes most human outbreaks from 2017, 2018 and 2022, and can be further divided into lineages A, A.1, A.1.1, and B.1.
In early May, 2022, MPXV cases were detected in the UK and Portugal, most of these with no known travel history outside the country of reporting. By the 9th June, 2022, 1240 MPXV cases had been confirmed in 33 countries (Global.health), mostly in European countries and the United States. Monkeypox typically presents clinically with fever, rash and swollen lymph nodes, and is transmitted to humans through close contact with an infected person or animal (WHOa, 2022). Available epidemiological and contact tracing data has revealed that currently, most cases are associated to men who have sex with men (MSM) (WHOb, 2022).
The first two MPXV cases from South America, from Argentina, were notified on the 27th of May, 2022 (Ministerio de Salud de la Nación Argentina 2022). Both cases reported travel to Spain 2 and 9 days before case notification. On the 3rd and 9th of June, 2022, two partial MPXV B.1 genomes from Argentina (942 bp) were deposited in GISAID as the first monkeypox sequences from Latin America.
As of the 8th of June, 2022, Brazil had 8 suspected monkeypox infections in the States of Santa Catarina, Ceará, Mato Grosso do Sul, Rio Grande do Sul, Rondônia and São Paulo (Brazilian Ministry of Health 2022).
Here we report a near complete genome of the first confirmed monkeypox case detected in Brazil. A skin swab of the lesions (vesicle and crust) was collected on June 7th, 2022 at Emilio Ribas Institute of Infectious Diseases, from a male patient, aged 41 years old, and with a recent travel history to Portugal and Spain.
Viral DNA was isolated from 200 μl of the recovered material using the QIAamp Viral DNA Mini Kit (Cat No. 51304, Qiagen, Germany) according to the manufacturer’s instructions and eluting in 60 μl of elution buffer. DNA was quantified using fluorimetry with the Qubit dsDNA High Sensitivity Assay (Cat No. Q32854, Life Technologies, USA) on the Qubit 3.0 instrument (Life Technologies, USA). Shotgun metagenomics was performed using 10 ng of the extracted DNA using the Rapid PCR Barcoding kit (SQK-RPB004) - Oxford Nanopore Technologies (ONT, UK), adapted from the Rapid SMART-9N protocol (Claro et al. 2021). PCR products were then purified using a 1:1 ratio of AMPure XP beads (Cat No. A63881, Beckman Coulter, UK) and quantified.
MinION libraries were prepared using an input of 50 ng per sample, pooled in an equimolar fashion followed by the rapid adapter ligation. The final libraries were loaded onto FLO-MIN106 flow cells on the MinION device (ONT, UK) and sequenced using MinKNOW 1.15.1 with the standard 48-hour run script. FASTQ files were demultiplexed and trimmed using Guppy V5.0.16, and the barcoded FASTQ files were aligned and mapped to the reference genome (GenBank accession no. MN648051) using minimap2 version 2.28.0 and converted to a sorted BAM file using SAMtools. NanoStat version 1.1.2 was used to compute the number of raw reads and minimum contig length to cover 50 percent of the genome (N50) of the aligned reads. Tablet 1.19.05.2827 was used for genome visualisation, and to compute the number of mapped reads, percentage of genome coverage, and coverage depth. Variants were detected with medaka_variants and the consensus sequence was built using medaka_consensus (ONT, UK). Genome regions with <20x coverage were masked.
Genome sequencing was performed upon sample receipt at the Institute Tropical Medicine, Sao Paulo, on the 7th June, and sequencing was completed in 18 hours, from DNA extraction to consensus sequence generation. Results were shared immediately upon generation and quality control with the patient’s healthcare facility and local public health authorities, and confirmed 24h later in a reference laboratory. Our sequencing run generated a total of 954,284 reads, with 784,000 reads with quality score > 8 used in the following analysis. The percentage of mapped reads was ~2%, with an average depth of 277.7x, covering 100% of the viral genome with at least 1 read, with an N50 of 4,493. Complete MPXV genome sequence BR0001 is also available under NCBI GenBank Accession Number ON751962.1. Consensus sequence and raw data can also be found in our project’s dedicated GitHub repository (GitHub - CADDE-CENTRE/Monkeypox: Monkeypox data).
To contextualise the novel monkeypox genome, we downloaded 102 whole-genome sequences from NCBI GenBank (Table S1). Of these, 81 were genomes from the ongoing MPXV B.1 multi-country outbreak. The dataset was aligned to the reference genome using MAFFT version 7·453 (Katoh & Standley 2013).
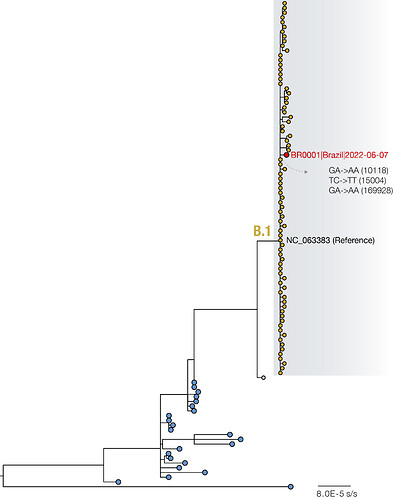
Figure. Maximum likelihood phylogeny with 103 whole genome sequences, including 102 available in NCBI GenBank up to the 9th June 2022 (see Table S1 for Accession Numbers used in this study). Highlighted genomes (yellow tips), including the SP01 described in this study (red tip), belong to the newly proposed B.1 lineage (Happi et al., 2022).
Maximum likelihood analysis (Minh et al. 2020) of near-complete and complete genome sequences revealed that the first MPXV case from Brazil grouped within the newly proposed B.1 lineage, and was closely related to sequences from Portugal, Germany, USA and Spain. We identified 3 unique SNPs compared to the NCBI reference genome (Accession number: NC_063383): GA->AA (10118), TC->TT (15004) and GA->AA (169928). Previous analysis suggests that G->A and C->T mutations as those observed in the new Brazilian genome are the effect of APOBEC3 deaminase editing (O’Toole and Rambaut, 2022).
Data Availability
Complete MPXV genome sequence BR0001 is also available under NCBI GenBank Accession Number ON751962.1. Acknowledgment table with Accession Numbers used for phylogenetic analysis shown in Figure, consensus sequence and raw data can be found in: GitHub - CADDE-CENTRE/Monkeypox: Monkeypox data.
Acknowledgments
This study was supported by the Medical Research Council-São Paulo Research Foundation (FAPESP) CADDE partnership award (MR/S0195/1 and FAPESP18/143890) (https://caddecentre.org). We acknowledge support from the Wellcome Trust and Royal Society (Sir Henry Dale Fellowship 204311/Z/16/Z), Bill & Melinda Gates Foundation (INV-034540 and INV-034652), the Wellcome ARTIC network (206298), the Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP 01.20.0029.000462/20, CNPq 404096/2020-4) and CNPq/MCTI funding 402794/2020-6.