First Dutch genome sequence of Monkeypox virus
Bas Oude Munnink, Marjan Boter, Babette Weller, Richard Molenkamp, Janette Rahamat-Langendoen, Reina Sikkema, Marion Koopmans
ErasmusMC, department of viroscience
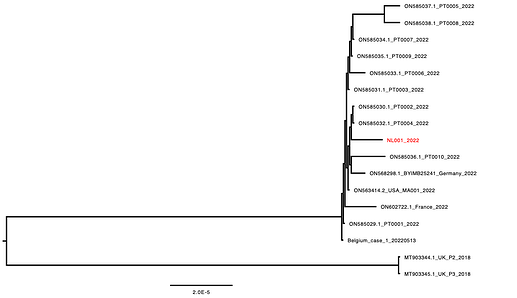
Here we announce the first Dutch genome sequence of the monkeypox virus associated with the currently ongoing multi-country outbreak. The virus clusters with the previously obtained sequences and only differs on two positions compared to the other sequences.
Nanopore sequencing was performed on clinical material after DNAse treatment. In short, DNA was extracted using Ampure Beads after which an end repair step was performed (Ultra II End-prep – NEBNext). Adapters were ligated, Ampure bead washes were performed twice and a random PCR was performed (following the SQK-PBK004 protocol from Nanopore).
Reads were mapped against a reference genome using Geneious after which a consensus was generated using a 10x coverage cut-off. The consensus sequence was manually curated consulting references sequences and the read alignment. Consensus sequences were aligned using Mafft after which the alignment was manually inspected and curated. A phylogenetic tree was made using IQ-TREE using the HKY+F model as best predicted model and the tree was visualized using FigTree.
The sequence has been submitted to GenBank but is also available here for early access:
NL001_2022.fasta.gz (58.6 KB)
Figure 1. Phylogenetic analysis of the sequences part of the multi-country 2022 monkeypox outbreak.