Cesare E.M. Gruber, Emanuela Giombini, Valentina Mazzotta, Serena Vita, Fabrizio Carletti, Martina Rueca, Giulia Gramigna, Alessandra D’Abramo, Stefania Cicalini, Daniele Lapa, Silvia Pittalis, Vincenzo Puro, Marco Rivano Capparuccia, Alessandra Marani, Lavinia Fabeni, Ornella Butera, Francesca Colavita, Silvia Meschi, Giulia Matusali, Eliana Specchiarello, Francesco Vairo, Francesco Vaia, Anna R. Garbuglia, Emanuele Nicastri, Andrea Antinori, Enrico Girardi for the INMI Monkeypox Group.
Monkeypox is a zoonotic virus that was first isolated in a monkey in 1958 in Denmark from Singapore [1]. The first human case of Monkeypox was identified in the Democratic Republic of Congo in 1970 [2] and, until now, the genetic analysis allowed to distinct two different clades related to the endemic area: West and Central Africa. Monkeypox symptoms are similar to those of smallpox infection, even if usually milder.
Monkeypox is a Poxviridae family member with a linear double-stranded DNA genome. The genome is over 197,000 nucleotide long with around 200 genes and two telomeres at the ends composed by inverted terminal repeats (ITR).
From 7 until 21 may 2022, 92 monkeypox cases have been reported in 12 non-endemic countries by WHO [3]. To date there have not been reported any fatal case.
Epidemiological investigations are ongoing, however, reported cases thus far have no established travel links to endemic areas. Based on currently available information, cases have mainly but not exclusively been identified amongst men who have sex with men (MSM) seeking care in primary care and sexual health clinics.
On May 18th, the first Italian case of the new outbreak was diagnosticated. The case was a 39-years-old Italian man returning from Gran Canaria Island on May 17th and was referred to our center. He has a HIV infection on effective antiretroviral treatment, and reported sexual condomless intercourses with different male partners. At admission, multiple asynchronous deep-seated and well-circumscribed lesions, with central umbilication, were present on the genital area with inguinal lymphadenopathy; a single lesion was present in the anterior and posterior thorax and in the left calf.
On May 26th, the first Italian near full genome was realised on GenBank (ON614676.1).
DNA extraction was performed from the scab sample of the first patient with Qiagen extraction kit. Next Generation Sequencing was performed using metagenomic approach. Library was prepared using Thermofisher Kit following manufacturer’s instructions and sequencing was performed on Ion Torrent GSS5 platform (ThermoFisher).
After quality filter, target reads owing to the poxvirus genome were de-novo assembled using SPAdes [4]. Major contigs were selected and aligned with BWA [5], to the Germany MPXV sequence recently published (ON568298.1), then the alignment was manually checked with Geneious Prime.
All reads were mapped to the obtained genome, and all variants were manually checked and eventually corrected. The assembled near to full genome of MPXV-INMI-Pt1 resulted of 190,280 nt, with a mean coverage of 159 reads. Annotation of 186 open reading frames was performed by transferring the annotation from the genome sequence MPXV-UK_P1 (MT903343.1) with Geneious Prime.
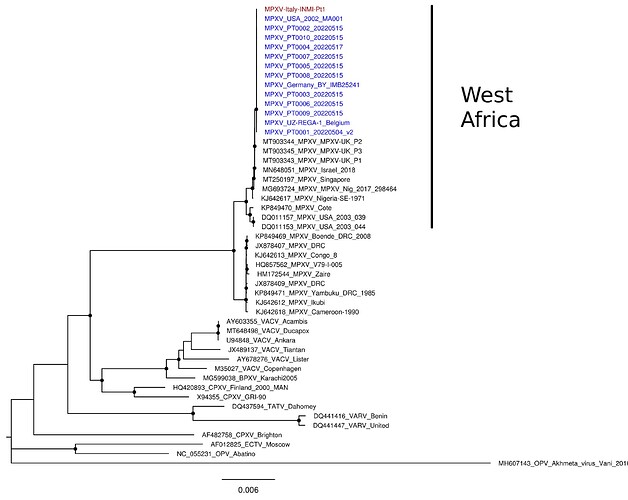
Whole genome sequences representative of the genus Orthopoxvirus were selected from GenBank and aligned with INMI-Pt1 and the available MPXV genomes from USA, Germany, Belgium and Portugal related to multi-country outbreak, using MAFFT [6].
Phylogenetic analysis was performed with IQ-TREE [7]: Kimura three parameters and FreeRate model with empirical base frequencies was selected with ModelFinder, and the best tree was found performing 5,000 bootstrap ultrafast replicates.
INMI Monkeypox Group: Alessandro Agresta, Francesco Baldini, Tommaso Ascoli Bartoli, Alessia Beccacece, Rita Bellagamba, Aurora Bettini, Nazario Bevilacqua, Marta Camici, Angela Corpolongo, Gabriella De Carli, Federico De Zottis, Francesca Faraglia, Massimo Francalancia, Concetta Maria Fusco, Roberta Gagliardini, Saba Gebremeskel, Maria Letizia Giancola, Elisabetta Grilli, Susanna Grisetti, Simone Lanini, Laura Emma Lapini, Gaetano Maffongelli, Andrea Mariano, Ilaria Mastrorosa, Claudia Minosse, Martina Moccione, Annalisa Mondi, Vanessa Mondillo, Nicoletta Orchi, Sandrine Ottou, Carmela Pinnetti, Silvia Rosati, Laura Scorzolini, Danilo Tacconi, Alessandra Vergori.
Refences:
1] Cho CT, Wenner HA. Monkeypox virus. Bacteriol Rev. 1973 Mar;37(1):1-18. doi: 10.1128/br.37.1.1-18.1973. PMID: 4349404; PMCID: PMC413801.
2] Ladnyj ID, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ. 1972;46(5):593-7. PMID: 4340218; PMCID: PMC2480792.
3] https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385.
4] Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012 May;19(5):455-77. doi: 10.1089/cmb.2012.0021. Epub 2012 Apr 16. PMID: 22506599; PMCID: PMC3342519.
5] Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25(14):1754-60. doi: 10.1093/bioinformatics/btp324. Epub 2009 May 18. PMID: 19451168; PMCID: PMC2705234.
6] Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002 Jul 15;30(14):3059-66. doi: 10.1093/nar/gkf436. PMID: 12136088; PMCID: PMC135756.
7] Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268-74. doi: 10.1093/molbev/msu300. Epub 2014 Nov 3. PMID: 25371430; PMCID: PMC4271533.