The Nigerian Centre for Disease Control (NCDC), the African Centre of Excellence for the Genomics of Infectious Disease (ACEGID) at Redeemer’s University, Ede, Nigeria (RUN), the Centre for Human Virology and Genomics (CHVG), Nigerian Institute of Medical Research (NIMR), the Centre for Human and Zoonotic Virology (CHAZVY), College of Medicine University of Lagos/ Lagos University Teaching Hospital (LUTH), the Lagos State Ministry of Health Bio-Safety Level 3 (BSL-3) Bio-Bank facility and partners report the first genome sequence of SARS-CoV-2 from Africa, from the first confirmed case of COVID-19 in Nigeria. This sequence is available at: [GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria].
On the 27th of February 2020, Nigeria confirmed its first case of COVID-19 at the Infectious Disease Hospital in Yaba, Lagos. The case was diagnosed by the Centre for Human and Zoonotic Virology (CHAZVY), College of Medicine University of Lagos/Lagos University Teaching Hospital (LUTH), part of the Laboratory Network of the NCDC. On the 1st of March 2020, a clinical specimen, specifically, a sputum specimen resuspended in 500µL of viral transport medium (VTM), was sent to ACEGID, Redeemer’s University, Nigeria, and Centre for Human Virology and Genomics, Nigerian Institute of Medical Research for sequencing and molecular characterization.
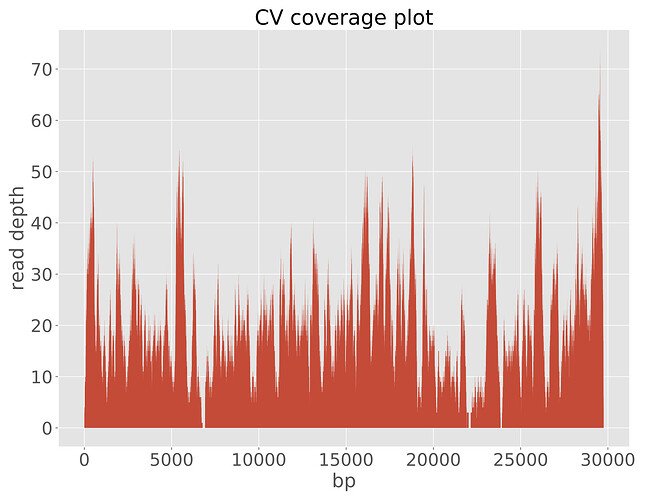
At ACEGID, viral RNA was extracted using the QiAmp viral RNA mini kit (Qiagen). RT-qPCR was repeated at ACEGID using the assay designed by the US CDC and confirmed the presence of SARS-CoV-2 viral RNA (Ct values of 28, 29, and 30 for N1, N2 and N3 viral gene targets, respectively), with no NTC amplification. Metagenomic sequencing libraries were prepared from total RNA as previously described (Matranga et al., 2016) and sequenced using one of the two Illumina MiSeqs in the sequencing platform of ACEGID. A full genome of SARS-CoV-2 was assembled with a length of 29,759 bp and mean coverage depth of 20X (Figure 1).
At the Centre for Human Virology and Genomics, NIMR, RT-qPCR was carried out using the assay designed by BGI China to confirm the presence of SARS-CoV-2 viral RNA with a Ct value of 26, targeting the high conservative region in 2019-nCoV genome. Sanger sequencing (ABI 3130 xl) using specific primer targeting the RdRp region was done. A partial sequence of SARS-CoV-2 (RdRp region) with a length of 486 bp (15321bp to 15806bp) was generated.
This report presents the first full genome sequence of SARS-CoV-2 from Africa to be released, with genomic data generated and assembled, and phylogenetic analysis performed by ACEGID and NIMR scientists. The turnaround time from sample receipt to sequence generation was 3 days (1st-4th of March 2020).
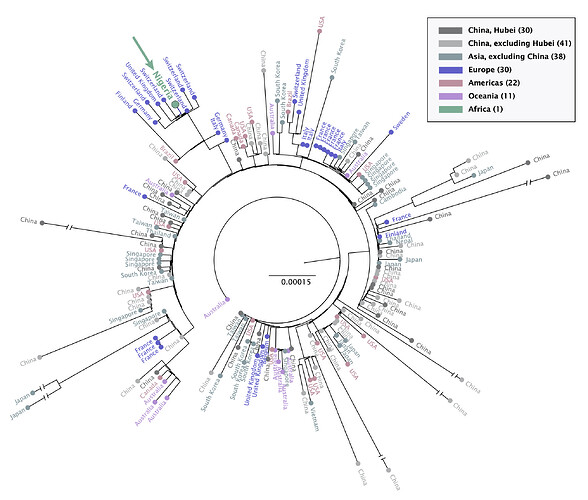
All HCoV whole genome sequences obtained from human hosts with geographical annotations were obtained from GISAID and aligned with the new genome from Nigeria. This genome clusters with a European clade, consistent with the known travel history of this case (Figure 2).
CV.coverage_plot.pdf (309.0 KB)
Figure 1: Illumina read coverage across coronavirus genome assembly from a patient in Nigeria.
20200305_COVID19_Nigeria_tree.pdf (205.3 KB)
Figure 2: Maximum likelihood tree of SARS-CoV-2 . These sequences were aligned using MAFFT v7.310 and tree reconstruction using IQTREE v1.6.1.
Data availability
All sequences are available at GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria
GISAID, NCBI GenBank, and NCBI SRA accession numbers will be shared when available. We would like to thank all the authors who have kindly deposited and shared genome data on GISAID. A table with genome sequence acknowledgments can be found at GitHub - acegid/CoV_Sequences: SARS-CoV-2 genome from Nigeria
Partners and Collaborators
Nigeria Centre for Disease Control (NCDC),
Abuja, Nigeria, @NCDCgov.
African Centre of Excellence for Genomics of Infectious Diseases (ACEGID),
Redeemer’s University, Ede, Osun State, @acegid_lab.
Centre for Human Virology and Genomics,
Nigerian Institute of Medical Research (NIMR).
Centre for Human and Zoonotic Virology (CHAZVY),
College of Medicine University of Lagos
Lagos University Teaching Hospital (LUTH).
Infectious Disease Hospital, Yaba, Lagos,
Lagos State Ministry of Health, Nigeria.
Redeemer’s University, Ede, Osun State (RUN).
Africa CDC, Addis Ababa, Ethiopia, @AfricaCDC.
Broad Institute and Harvard University, Cambridge, MA, USA.
Beth Israel Deaconess Medical Center, Boston, MA, USA.
China Centre for Disease Control, Beijing, China.
Disclaimer and contact information
Please note that these analyses are based on work in progress and should be considered preliminary. Our analyses of this data are ongoing and a publication communicating our findings on these and other published genomes is in preparation. If you wish to use this data please contact:
Chikwe Ihekweazu, M.P.H, F.F.P.H
Director General, Nigerian Centre for Disease Control (NCDC)
Abuja, Nigeria
Email: [email protected]
Website: [https://ncdc.gov.ng/]
Twitter: @Chikwe_I
Christian Happi, PhD
Professor of Molecular Biology and Genomics, Redeemer’s University, Ede, Osun State, Nigeria
Director, African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
E-mail: [email protected]
Website: www.acegid.org
Twitter: @christian_happi
Sunday Omilabu, PhD
Professor & Director, Centre for Human and Zoonotic Virology (CHAZVY),
College of Medicine University of Lagos
Lagos University Teaching Hospital (LUTH)
Email: [[email protected]]
Website: https://www.luth.org.ng/
Twitter: @AremuOmilabu
Babatunde Lawal Salako, MBBS, FWACP, FRCP (Edin), FRCP (Lond), MNIM, FAS
Director General, Nigerian Institute of Medical Research (NIMR)
Yaba, Lagos
Email: [email protected]
Website: https://www.nimr.gov.ng/
Twitter: @LawalSalako
Akin Abayomi, MBBS, MRCP, FCPATH
Professor & Honorable Commissioner for Health, Lagos State
Email: [email protected]
Website: https://www.health.lagosstate.gov.ng
Twitter: @ProfAkinAbayomi
Paul Eniola Oluniyi, (PhD in view)
African Center of Excellence for Genomics of Infectious Diseases (ACEGID)
Redeemer’s University, Ede, Osun State, Nigeria
E-mail: [email protected]
Website: www.acegid.org
Twitter: @pauloluniyi