Electron microscopy and a sequence-independent, single-primer amplification (SISPA) viromics approach for monkeypox virus genome determination
Nicola Fletcher 1,2*,†, Gabriel Gonzalez 3,4†, Luke Meredith 5†, Tiina O’Neill 2, Kevin Purves 1,2, Daniel Hare 3, Brian Keogan 3, Charlene Bennett 3, Joanne Moran 3, Sophie O’Reilly 6, Lili Gu 6, Matthew Angeliadis 6, Marion Pouget 6, Dimitri Scholz 2, Virginie Gautier 6, Patrick Mallon 6,7, Stephen Gordon 1,2, Jonathan Dean 3, Jeff Connell 3, Michael Carr 3,4, Cillian De Gascun 3*.
- UCD School of Veterinary Medicine, University College Dublin, Belfield, Dublin, Ireland.
- UCD Conway Institute, University College Dublin, Belfield, Dublin, Ireland.
- National Virus Reference Laboratory (NVRL), School of Medicine, University College Dublin, Belfield, Dublin, Ireland.
- International Collaboration Unit, International Center for Zoonosis Control, Hokkaido University, N20 W10 Kita-ku, Sapporo 001-0020, Japan.
- Department of Pathology, Division of Virology, University of Cambridge, Cambridge, United Kingdom.
- Centre for Experimental Pathogen Host Research (CEPHR), University College Dublin, Dublin, Ireland.
- Department of Infectious Diseases, St Vincent’s University Hospital, Elm Park, Dublin, Ireland.
*Correspondence to: [email protected] and [email protected].
†These authors contributed equally.
Monkeypox is a viral zoonosis endemic in West and Central Africa and presents clinically in humans as a smallpox-like illness (Bunge et al., 2022). Monkeypox virus (MPXV) is an enveloped virus with a particle size of ca. 300×240 nm containing a double-stranded DNA genome of ~197 kb taxonomically classified in the family Poxviridae, genus Orthopoxvirus (Weaver and Isaacs, 2008). There is increasing public health concern about the geographical spread and re-emergence of MPXV arising from the recent multi-country outbreaks, with more than 47 206 cases reported in 92 non-endemic countries since the first case was laboratory confirmed in the United Kingdom on the 7th of May 2022 (updated August 28th 2022 (CDC, 2022)). The MPXV genome reported (GenBank ON872184) here was from a laboratory-confirmed Irish case (Orthopox group RNA and MPXV-specific RNA detection by real-time PCR) identified in June 2022. Transmission electron microscopy (TEM) of a lesion swab sample collected visualised multiple characteristic brick-shaped poxvirus virions with lattice-like surface detail (Fig. 1). We performed a sequence-independent, single-primer amplification (SISPA) viromics approach to achieve a non-selective (unbiased) amplification of viral RNA transcripts and here report horizontal genome coverage over 90% with an average vertical coverage (depth) between 9x and 21x. Phylogenetic analysis with 35 publicly available international MPXV genome sequences from multiple countries (UK, Israel, Nigeria, USA, Singapore and Portugal) sampled between 2017-2019 and 2022 (Happi et al.), demonstrated high sequence similarity to other cases circulating in the 2022 outbreak in Europe and USA clustered together in clade 3 (Happi et al.); with the Irish MPXV genome further clustered in the B.1 lineage (Fig. 2).
Methods
Sample collection and DNA extraction. Following lysis and inactivation in a Containment Level 3 (CL3) facility with a viral transport medium (VTM) to guanidinium isothiocyanate lysis buffer mixed in a ratio of 1:2.5, total nucleic acids were extracted on the Roche MagNA Pure 96 platform as follows: 200 µL of swab material in VTM was added to 500 µL Roche lysis buffer, and extracted on the Roche MagNA Pure 96 system (input volume 450 µL and eluted in 100 µL). Alternatively, DNA from 200 µL of material in VTM was extracted using the Qiagen QIAamp DNA Mini kit, according to the manufacturer’s instructions.
Molecular Diagnostics. Total nucleic acids were extracted from clinical specimens (swabs in VTM) on the Roche MagNA Pure 96 automated platform for routine diagnostics after lysis in a CL3 laboratory for molecular diagnostics. Real-time PCR was performed on the ABI7500 SDS platform employing the Orthopoxvirus RT-PCR kit (Altona) as per the manufacturer’s instructions, or the previously described assays for generic MPXV and West African clade (Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA - PubMed).
Transmission electron microscopy (TEM). One sample (sample Z22IRL00094) was filtered through a 0.45 µm filter and 40 µL filtrate fixed in an equal volume of 5% glutaraldehyde for 1 h at room temperature. Sample was placed on formvar-coated copper 400 electron microscopy grids for 5 minutes; excess supernatant was removed and the grids were stained with 1 % uranyl acetate. Grids were visualized using a Tecnai 12 transmission electron microscope.
Sequence-Independent, Single-Primer Amplification (SISPA) amplification. DNA was amplified using a SISPA approach adapted from Kafetzopoulou et al, 2018. Primer A - 5’-GTTTCCCACTGGAGGATA-(N9)-3’ was annealed to extracted DNA by incubation at 65°C for 5 minutes followed by 4°C for 5 minutes. First strand cDNA synthesis was performed using Superscript IV, and incubated at 50°C for 10 min followed by addition of Sequenase V2.0 DNA polymerase, according to manufacturer’s instructions, and incubated at 37°C for 8 minutes. DNA amplification was performed using AccuTaq LA (Sigma, Poole, United Kingdom), in which 5 μL of DNA and 1 μL (40 pmol/μL) of Primer B (5′-GTTTCCCACTGGAGGATA-3′) were added to a 50 μL reaction, according to the manufacturer’s instructions. PCR conditions were as follows: 98°C for 30s, followed by 30 cycles of 94°C for 15 s, 50°C for 20 s, and 68°C for 5 min, and a final step of 68°C for 10 min. Amplified cDNA was purified using a 1:1 ratio of AMPure XP beads (Beckman Coulter, Brea, California (CA)) and quantified using the Qubit High Sensitivity dsDNA kit (Thermo Fisher, Waltham, US). A no template control was included in each sequencing run.
MinION library preparation and sequencing. Individual SISPA-amplified samples were barcoded using the rapid barcoding kit SQK-RBK004 (Oxford Nanopore Technologies, ONT), then run pooled and run on a R9.4.1 flow cell, employing a 48 h run script with high base-calling accuracy enabled.
Sequence Assembly and Analysis The data generated was basecalled and demultiplexed with Guppy v5.1.13 (ONT) using the high accuracy model and for the barcode kit stated above. The sequence reads were trimmed to remove adapters and barcode sequences with Porechop v.0.3.2pre (GitHub - rrwick/Porechop: adapter trimmer for Oxford Nanopore reads). The MPXV reads were aligned and mapped with BWA (Li and Durbin, 2009) against the reference genome with accession number ON563414. Medaka (GitHub - nanoporetech/medaka: Sequence correction provided by ONT Research) was used to polish the assembled genomes. The assembled sequences were multiple-sequence aligned with publicly-available sequences in the NCBI database (www.ncbi.nlm.nih.gov) using the program MAFFT (Yamada, Tomii, and Katoh, 2016). A phylogenetic tree was inferred using RAxML (Stamatakis, 2014) with support for the branches estimated using the bootstrap method with 200 repetitions.
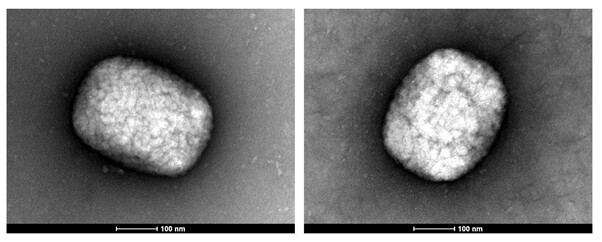
Figure 1. TEM images of MPXV particles identified in Z22IRL00094 illustrating characteristic brick-shaped poxvirus morphology. The scale bar is shown at the bottom of both images.
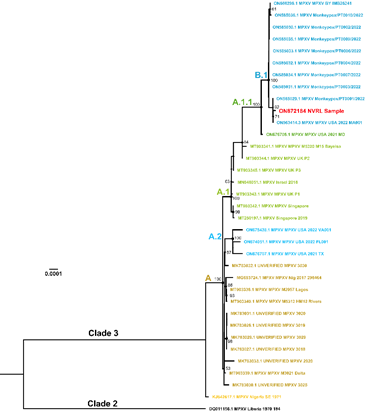
Figure 2. Maximum likelihood phylogenetic tree obtained from the MPXV alignment. Irish partial MPXV genome sequence (GenBank accession number ON872184) is coloured in red. The clades are annotated according to Happi et al. (2022).
References
Bunge, E.M., Hoet, B., Chen, L., Lienert, F., Weidenthaler, H., Baer, L.R. and Steffen, R., 2022. The changing epidemiology of human monkeypox-A potential threat? A systematic review. PLoS Negl Trop Dis 16, e0010141.
CDC. 2022. 2022 Monkeypox Outbreak Global Map. 2022 Mpox Outbreak Global Map | Mpox | Poxvirus | CDC.
Happi, C., Adetifa, I., Mbala, P., Njouom, R., Nakoune, E., Happi, A., Ndodo, N., Ayansola, O., Mboowa, G. and Bedford, T., Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus.
Li, H. and Durbin, R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-60.
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312-3.
Weaver, J.R. and Isaacs, S.N., 2008. Monkeypox virus and insights into its immunomodulatory proteins. Immunol Rev 225, 96-113.
Yamada, K.D., Tomii, K. and Katoh, K., 2016. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 32, 3246-3251.