On 29 July 2024, the Kenyan Ministry of Health confirmed a case of mpox in Taita Taveta County which borders Tanzania (Mpox – African Region). The reported case was from a 42-year-old male, a long-distance truck driver who had travelled from Kampala, Uganda to Mombasa, Kenya and was enroute to Rwanda at the time of detection. The date of onset of symptoms was 3 July 2024 and the case was detected and isolated by the Kenyan Ministry of Health officials on 22 July 2024 after presenting with classic symptoms of mpox infection. Here, we report the rapid application of unbiased metagenomic next generation sequencing (mNGS) to reconstruct the genome sequence of this mpox case in Kenya. The protocol under which the sample was obtained and processed for qPCR and sequencing is approved by the Kenya Medical Research Institute’s Scientific Ethics and Review Unit (SERU) under study number; SSC3035.
DNA extraction was carried out on the clinical sample (lesions and vesicle swabs) using Biocomma DNA/RNA virus purification kit. MPXV was detected on the extracted DNA using the Liferiver Real Time PCR Kit and subsequently subjected to unbiased mNGS. Libraries were prepared using NEBNext Ultra II DNA library prep kit for Illumina (NEB, UK), and sequenced on an Illumina iSeq 100 to generate 2x146bp paired-end reads. Approximately 6.3 million total reads were obtained and we recovered a near complete MPXV genome with an average depth of coverage of ~20.3x.
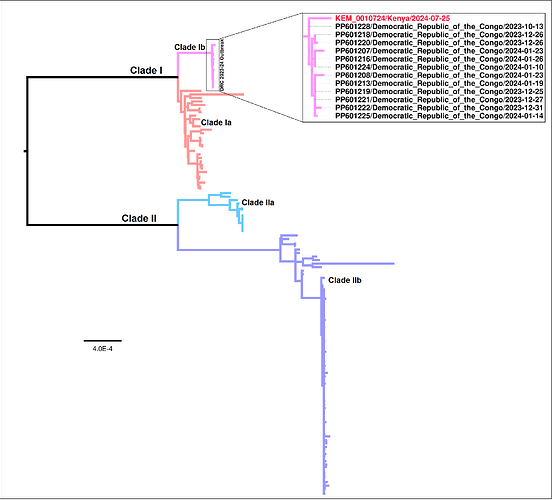
Our findings show that the Kenyan case clusters together with clade Ib MPXV strains, associated with the sustained outbreak in the DRC (Figure 1) (Vakaniaki et al., 2024). Clade Ib lineage has been associated with continuing geographical expansion of the virus to previously unaffected areas, high incidence of the disease as well as high case fatality (CFR 4.9-6.7%). Similar to other clade Ib strains, the Kenyan strain carries APOBEC3-type mutations which highlights the need for surveillance to curtail any potential expansion of this MPXV strain.
Figure1: Maximum Likelihood phylogeny based on complete genomes belonging to the two major clades of MPXV. The DRC outbreak clade has-been expanded with the sequenced strain from Kenya indicated with a red tip label.
In conclusion, we report the genome sequence and confirmation of clade Ib strain in Kenya. Our analysis show that this case carries APOBEC3-type mutations, similar to other clade Ib strains from the DRC. These findings highlight the need for continued surveillance to curtail any potential expansion of this MPXV strain in Kenya and the region.
Data Availability
The sequencing data generated can be found in NCBI BioProject PRJNA1147890 and the genome sequence reported in the study has been submitted to Genbank under accession number PQ178862.
Authors and Funding:
Sequencing and data analysis were made possible through the support of KEMRI, funding from Bill and Melinda Gates Foundation and Chan Zuckerberg Initiative and also the public health team that identified the suspect case and facilitated the collection and subsequent isolation of the patient until confirmation of this case.
The following individuals are leading this work:
Ministry of Health – Kenya: Emmanuel Okunga, Daniel Langat and Patrick Amoth.
Kenya Medical Research Institute (KEMRI): Konongoi Limbaso, Victor Ofula, Samoel Khamadi and Elijah Songok.
Chan Zuckerberg Biohub (US): Genay Pilarowski and Paul Oluniyi.