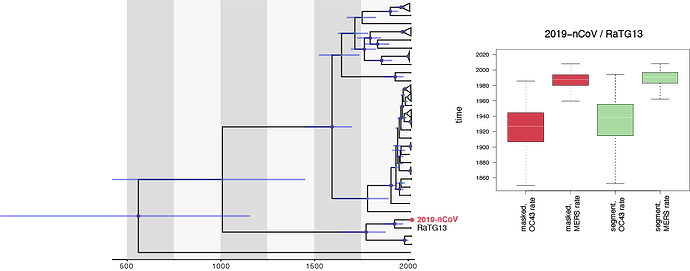
Following on from the recombination analyses discussed in a different thread, specifically the analyses by David Robertson and Maciek Boni, we performed similar dating analyses using a full genome alignment were recombination was masked and an alignment that concatenates segments with consistent phylogenetic signal (11.5 kb). As calibration we used a rate distribution based on OC43 (0.00024 [0.00019,0.00029] subst./sites/yr) and MERS (0.00078 [0.00063,0.00092] subst./sites/yr):
Using the OC43 rate, we arrive at a mean estimate for 2019-nCoV/RaTG13 split of about 1925-1930 for both data sets, whereas the MERS rate results in a younger estimate (a mean age of about 1988). The differences with Andrew’s estimates and the variability of all these estimates appears to be almost entirely due to the choice in the rates that are used, raising the question which rates estimates are more appropriate for this.