Context
On April 23, 2022, the Minister of Public Health, Hygiene, and Prevention announced a new case of Ebola virus disease (EVD) occurring in Mbandaka health zone in Equateur Province, Democratic Republic of the Congo (DRC). EVD outbreaks occurred previously in Equateur Province in 2018 and 2020.
The 31-year-old male, a student from Boende in Tshuapa Province, returned to Mbandaka, Equateur Province on February 25, 2022. He was vaccinated against Ebola virus in 2020 and is not known to be an EVD survivor. Symptom onset occurred on April 05, 2022 with fever and headache, which were treated at home. He was admitted to the Bongisa Libota Health Center in Mbandaka on April 16, where he was hospitalized for three days, before being transferred on April 19 to the Wangata General Reference Hospital in Mbandaka. On April 21, signs of haemorrhagic fever appeared, and the patient was placed in isolation at the Wangata Ebola treatment center as a suspected EVD patient but died the same day.
Blood and swab samples were taken, which tested positive for Ebola virus at the Mbandaka provincial laboratory on April 21. The two samples were sent to the Institut National Recherche Biomedicale (INRB) in Kinshasa where Ebola virus was confirmed by GeneXpert on April 23.
Sequencing
The blood and oral swab samples were sequenced at the Pathogen Genomics Laboratory at INRB-Kinshasa on an Oxford Nanopore Technologies GridION. Sequencing libraries were prepared using an amplicon sequencing approach (Mbala-Kingebeni, 2021; Grubaugh et al., 2019; Quick et al. 2017), generating identical genomes with 98.3% and 96.7% genome coverage.
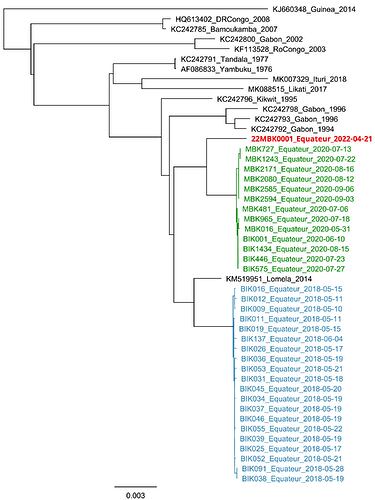
Figure 1. Maximum likelihood phylogenetic tree of all EVD outbreaks, midpoint rooted, including genome sequences from the Equateur 2018 (blue), Equateur 2020 (green), and new Equateur 2022 (red) outbreaks.
The two genomes were identical and the genome with highest coverage (22MBK0001) was aligned with genomes from prior sequenced Ebola virus disease outbreaks using MAFFT (Katoh, 2013). A phylogenetic tree was inferred using RAxML (Stamatakis, 2014; Figure 1). The new genome was most closely related to the Equateur 2020 outbreak but fell outside the outbreak clade, indicating that this case represents a new spillover event from the host reservoir and is not directly linked to the Equateur 2018 or Equateur 2020 EVD outbreaks. Further epidemiologic investigations are ongoing.
Data availability
The genome sequence and figure are available here
Partners and collaborators
Placide Mbala-Kingebeni (INRB, University of Kinshasa)
Jean-Jacques Muyembe (INRB, University of Kinshasa)
Steve Ahuka-Mundeke (INRB, University of Kinshasa)
Hugo Kavunga (INRB)
Daniel Mukadi (INRB, University of Kinshasa)
Elisabeth Pukuta (INRB)
Eddy Kinganda Lusamaki (INRB, University of Kinshasa)
Amuri Aziza (INRB, University of Kinshasa)
Jean Claude Makangara (INRB, University of Kinshasa)
Emmanuel Lokilo (INRB)
Franck Edidi (INRB, University of Kinshasa)
Junior Bula Bula (INRB, University of Kinshasa)
Raphael Lumembe (INRB, University of Kinshasa)
Gabriel Kabamba (INRB, University of Kinshasa)
Fabrice Mambu (INRB-Goma)
Joel Montgomery (CDC Atlanta)
Peter Fonjungo (CDC Atlanta)
Norbert Soke (CDC Atlanta)
Jacques Likofata (WHO DRC)
Vital Mondonge (WHO DRC)
Gervais Folefack (WHO DRC)
Prosper Djiguimde (WHO DRC)
John Otshudiema (WHO DRC)
Deby Mukendi (WHO DRC)
Dieudonné Mwamba (DGLM DRC)
John Kombe (DGLM DRC)
Sofonias Tessema (Africa CDC)
Didi Bofaka (Laboratoire Provincial de Mbandaka)
Andrew Rambaut (University of Edinburgh)
Mike Wiley (University of Nebraska Medical Center [UNMC], PraesensBIO)
Catherine Pratt (UNMC)
We gratefully acknowledge the Africa Pathogen Genomics Initiative - Africa CDC for support for equipment and reagents.
Statement on continuing work and analyses prior to publication
This genome is being shared pre-publication. Please note that this data is based on work in progress and should be considered preliminary. Our analyses of these data are ongoing and a publication communicating our findings is in preparation. If you intend to use these sequences prior to our publication, please communicate with Dr. Placide Mbala for coordination.
References
Grubaugh ND, Gangavarapu K, Quick J, et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 2019;20:8. 10.1186/s13059-018-1618-7 4 7
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772-80. 10.1093/molbev/mst010
Mbala-Kingebeni P, Pratt C, Mutafali-Ruffin M, et al. Ebola Virus Transmission Initiated by Relapse of Systemic Ebola Virus Disease. N Engl J Med 2021; 384:1240-1247 10.1056/NEJMoa2024670 12
Quick, J., Grubaugh, N., Pullan, S. et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 2017;12:1261–1276. 10.1038/nprot.2017.066 2
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312-3. 10.1093/bioinformatics/btu033