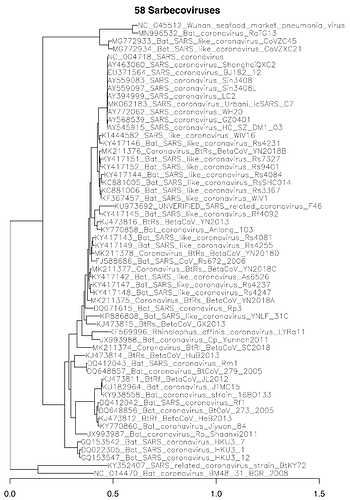
We have created an alignment of 58 Sarbecovirus complete genomes with SARS-CoV-2 reference having phylogenetic branch length suitable for analysis of conservation by programs like phyloP, PhyloCSF, and FRESCo. The alignment contains one strain of SARS-CoV-2, several strains of SARS-CoV, and, guessing from the names, the rest are bat viruses. Branch length of the tree is approximately 3 substitutions at 4-way synonymous sites, which is comparable with the 29-mammals and 12-flies alignments that have been used for that purpose in human and Drosophila, respectively.
SarbecovirusGenomes.RAxML.Mapped.nh.pdf (136.3 KB)
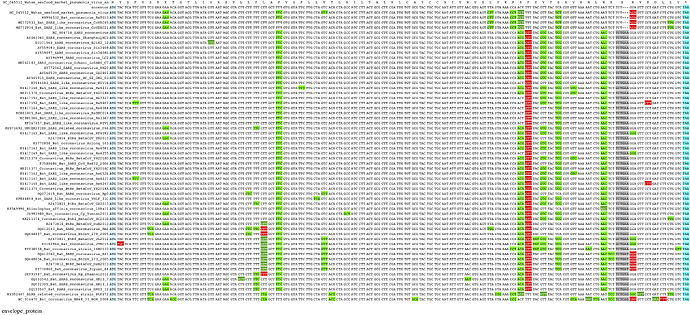
The alignments for the annotated SARS-CoV-2 genes can be viewed in CodAlignView using the following URLs:
ORF3a_protein
ORF6_protein
envelope_protein
orf1ab_polyprotein
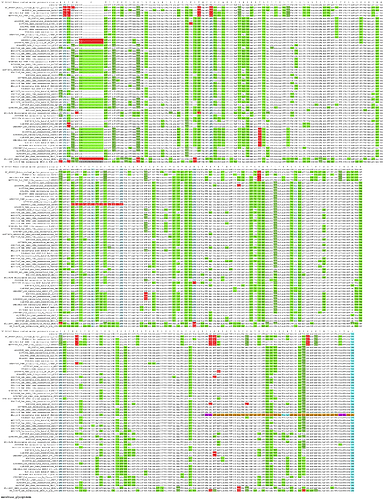
membrane_glycoprotein
ORF8_protein
nucleocapsid_phosphoprotein
surface_glycoprotein
ORF7b
ORF7a_protein
orf1a_polyprotein
ORF10_protein
The alignments are color coded to distinguish synonymous substitutions (light green) from conservative (dark green) and radical (red) amino acid changes. Frame-shifted regions are in orange.
The number of substitutions at 4-way synonymous sites varies widely by gene:
ORF3a_protein 3.7
ORF6_protein 2.1
envelope_protein 1.2
orf1ab_polyprotein 3.5
membrane_glycoprotein 3.3
ORF8_protein No 4-way sites with full branch length.
nucleocapsid_phosphoprotein 2.2
surface_glycoprotein 6.3
ORF7b 1.8
ORF7a_protein 3.6
orf1a_polyprotein 3.8
ORF10_protein No 4-way sites with full branch length.
There is considerable synonymous constraint throughout the length of the envelope protein (which is only 76 codons long), suggesting that there is some purifying selection in this region in addition to the purifying selection on the amino acid sequence. We do not know what might be causing this purifying selection, but it cannot be caused by some cryptic ORF in an overlapping reading frame because there are several internal stop codons in the other frames.
Here’s are the alignments for the envelope protein and, for comparison, the membrane glycoprotein:
The genomes were downloaded from NCBI and aligned using clustalo. The tree was created using RAxML with the GTRCATX model. Alignments in MAF and Fasta formats and the tree in Newick format are attached. The 58 accessions are: NC_045512, MN996532, MG772933, MG772934, NC_004718, AY463060, EU371564, AY559083, AY559097, AY394999, MK062183, AY772062, AY568539, AY545915, KT444582, KY417146, MK211376, KY417151, KY417152, KY417144, KC881005, KC881006, KF367457, KU973692, KY417145, KJ473816, KY770858, KY417143, KY417149, MK211378, FJ588686, MK211377, KY417142, KY417147, KY417148, MK211375, DQ071615, KP886808, KJ473815, KF569996, JX993988, MK211374, KJ473814, DQ412043, DQ648857, KJ473811, KU182964, KY938558, DQ412042, DQ648856, KJ473812, KY770860, JX993987, GQ153542, DQ022305, GQ153547, KY352407, and NC_014470.
SarbecovirusGenomes.RAxML.nh.gz (1.5 KB) SarbecovirusGenomes.fa.gz (286.3 KB) NC_045512.2.maf.gz (414.6 KB)