A brief update to this post: Initial observations about putative APOBEC3 deaminase editing driving short-term evolution of MPXV since 2017
Áine O’Toole & Andrew Rambaut
Institute of Ecology and Evolution
University of Edinburgh
2022-06-05
Three genome sequences from the USA (two from 2022, one from 2021) have produced evidence of a new lineage clustering amongst the 2017 genomes. And another one from USA from 2021 splitting the long branch to the global 2022 lineage.
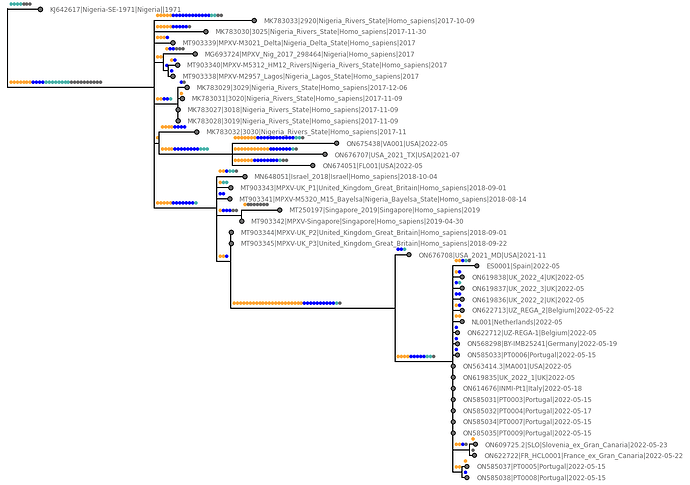
Figure 1 | Orange dots denote
GA→AAmutations, blue dots denoteTC→TTmutations (both predicted APOBEC3 dinucleotide mutations), cyan dots are otherG→AorC→Tmutations, and grey dots are other mutations.
All of these genomes exhibit the same pattern of having a predominance of likely APOBEC3 deaminase induced mutations.
A substantial number of the 2022 genomes have unique or shared APOBEC3 mutations confirming this mechanism as being the likely source of most genetic single nucleotide variation in humans within this lineage of MPXV.
If these APOBEC3 edits are specifically indicative of replication in humans as opposed to another host species then this would confirm this entire clade to be representative of the emergence of a human epidemic by 2017.
New USA genomes:
| accession | name | reference |
|---|---|---|
| ON676708 | MPXV_USA_2021_MD | Gigante et al, 2022, CDC |
| ON674051 | MPXV_USA_2022_FL001 | Gigante et al, 2022, CDC |
| ON676707 | MPXV_USA_2021_TX | Gigante et al, 2022, CDC |
| ON675438 | MPXV_USA_2022_VA001 | Gigante et al, 2022, CDC |