28 Jan 2020
Emma Hodcroft, Biozentrum, University of Basel, Switzerland
emma.hodcroft(at)unibas.ch
In order to provide context and background to the 2019 novel Coronavirus (nCoV-2019), we have prepared a Nextstrain/augur/auspice pipeline which automates full-genome, human-focused builds of Betacoronavirus 1, Human coronavirus 229E, and SARS-CoV, using data from ViPR.
Analyses
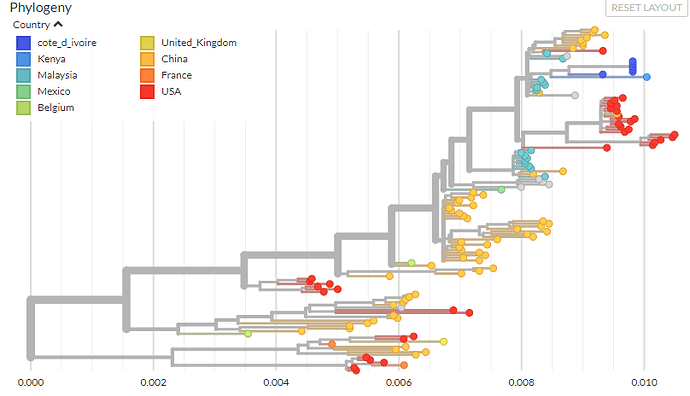
Betacoronavirus 1
The phylogenetic analysis of the Betacoronavirus 1 sequences is filtered to human and chimpanzee infections, and thus contains only Human coronavirus OC43 sequences.
HCoV-OC43 is distributed worldwide, considered a ‘seasonal’ coronavirus, and is one of the viruses responsible for the common cold.
Sequences date from between 1997 and 2019 and cover 9 countries, and the estimated mutation rate is between 2-3 x10-4 subs/site/yr.
The build can be viewed live here:
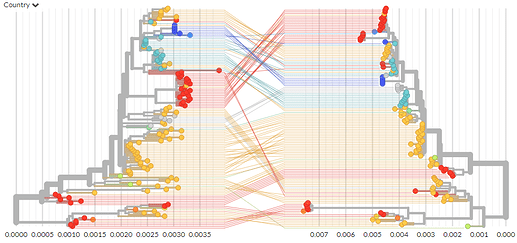
A fairly rough tanglegram comparing the first ~20,000bp to the last ~10,000bp can be viewed here:
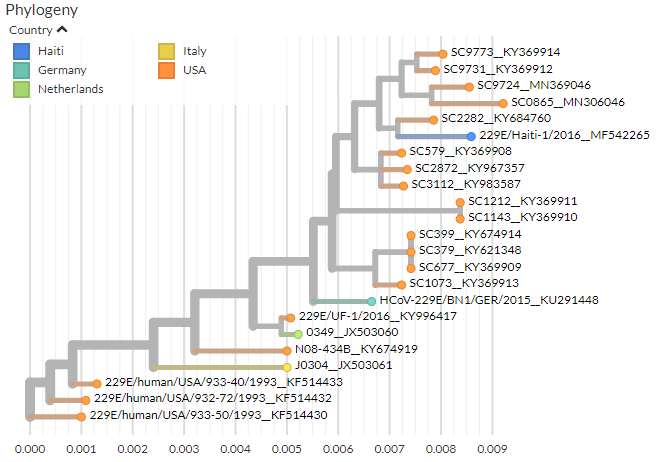
229E
Human coronavirus 229E can be found in animals, including camels and bats, but the phylogenetic analysis here is filtered to only human samples.
Like HCoV-OC43, HCoV-229E is distributed worldwide and responsible for the common cold.
Sequences date from 1993 to 2019 and cover 5 countries, with an estimated mutation rate of 2-3 x10-4 subs/site/yr.
The build can be viewed live here:
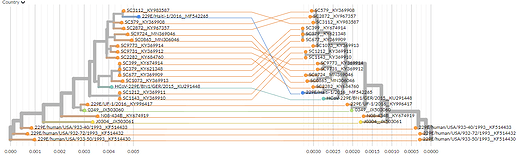
A fairly rough tanglegram comparing the first ~20,000bp to the last ~10,000bp can be viewed here:
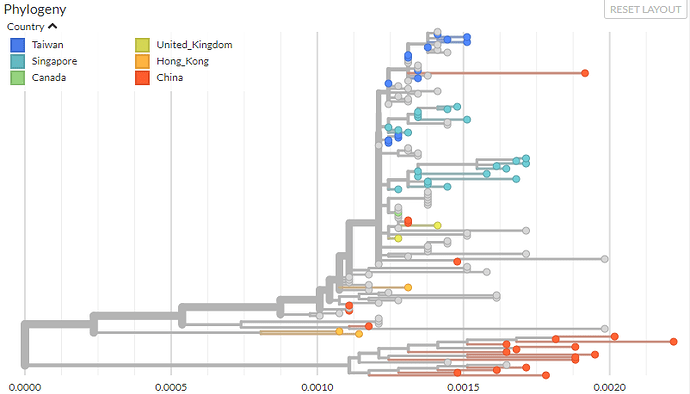
SARS
The Severe acute respiratory syndrome-related coronavirus phylogenetic analysis was filtered to exclude samples from bats, as these are more divergent*. The majority of the remaining samples are from palm civets and humans.
*nCoV-2019 sits among these bat samples.
SARS-CoV was responsible for an outbreak of severe respiratory illness in Asia in 2002-2003 (with secondary cases worldwide). There have been no cases of SARS in humans since 2004.
Sequences range from 2002-2004 in 6 countries, with a mutation rate estimated at ~3.6 x10-4 subs/site/yr.
The build can be viewed live here:
About the builds
All code and data, plus information about how to run the builds, plus the assumptions made for each run and some details on filtering and exclusions, can be found at github.com/nextstrain/cov.
The builds are filtered to be human-focused, and can be easily updated if/when new sequences are available on ViPR, by automatic detection of which sequences are new since the last run. Only these will be downloaded from Genbank and aligned.