Joana Isidro1, Vítor Borges1, Miguel Pinto1, Rita Ferreira1, Daniel Sobral1, Alexandra Nunes1, João Dourado Santos1, Maria José Borrego3, Sofia Núncio2, Ana Pelerito2, Rita Cordeiro2, João Paulo Gomes1,*.
1 Bioinformatics Unit, Department of Infectious Diseases, National Institute of Health Doutor Ricardo Jorge (INSA), Lisbon, Portugal
2 Emergency and Biopreparedness Unit, Department of Infectious Diseases, National Institute of Health Doutor Ricardo Jorge (INSA), Lisbon, Portugal
3 National Reference Laboratory of Sexually Transmitted Infections, Department of Infectious Diseases, National Institute of Health Doutor Ricardo Jorge (INSA), Lisbon, Portugal
contact: [email protected]
Monkeypox is a rare zoonotic disease that is caused by the monkeypox virus, belonging to the Orthopoxvirus genus, which also includes the variola virus (causes smallpox) (1). Although the natural reservoir of monkeypox remains unknown, rodents and non-human primates may harbor the virus and infect people. It was firstly identified in 1958 when two outbreaks of a pox-like disease occurred in monkeys kept for research purposes (2) and in 1970 in humans in the Democratic Republic of the Congo (3). Monkeypox is now endemic in some Central and West African countries (1), being rarely reported outside Africa. The virus is transmitted from one person to another by close contact with lesions, body fluids, respiratory droplets and contaminated materials (Monkeypox).
In May 2022, multiple cases of monkeypox were identified in several non-endemic countries, such as Portugal, the United Kingdom, Spain, Sweden, Belgium and the United States of America. Studies are currently underway to further understand the introduction and rapid dissemination of monkeypox in these countries. In this regard, the determination of the genome sequence of the virus causing these infections will certainly contribute to better understand the epidemiology, sources of infection, and transmission patterns.
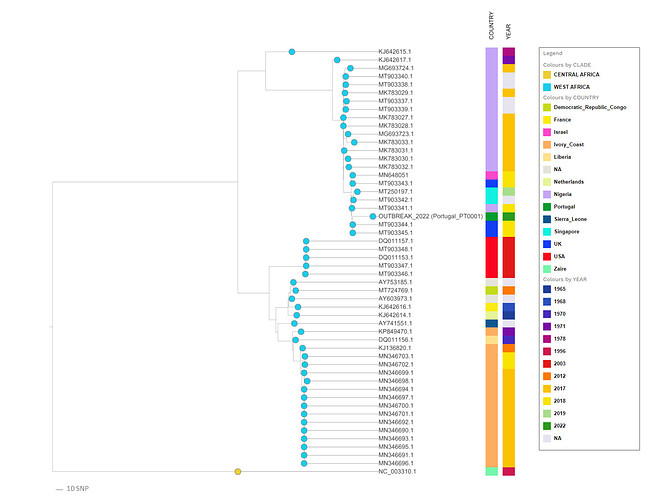
On May 19th, Portugal had already registered more than 20 laboratory confirmed cases of monkeypox and several samples were already subjected to laboratory procedures towards the determination of the virus genome sequence. Here we report the release of the first draft genome sequence of the monkeypox virus associated with this major outbreak affecting multiple countries worldwide. This sequence (Monkeypox_PT0001_2022.zip (52.1 KB)) was obtained from a swab collected on May 4th from skin lesions from a male patient. A first rapid phylogenetic analysis of the draft genome (Figure 1) indicates that the 2022 virus belongs to the West African clade and is most closely related to viruses associated with the exportation of monkeypox virus from Nigeria to several countries in 2018 and 2019, namely the United Kingdom, Israel and Singapore (4, 5). This preliminary data and analysis will be soon updated upon the release of new genome data, which will be important to elucidate the origin and international spread of the currently circulating virus.
Figure 1. Draft phylogenetic analysis based on a core alignment (955 variant positions in a 137 668 bp alignment) including 52 Monkeypox viral sequences.
Brief description of the methods
DNA was extracted from skin exudate samples using the QIAamp DNA Blood kit (Qiagen), prior to library preparation using the Rapid Barcoding Sequencing kit (SQK-RBK004) and shotgun metagenomics sequencing on an Oxford Nanopore MinION apparatus. Mean depth of coverage throughout the genome was around 7-fold. Reads were mapped to a closely related reference genome sequences (MN648051.1) using the INSaFLU online platform (https://insaflu.insa.pt/) and genome sequence was manually inspected to validate variant positions. The genome sequences will be further curated (namely to refine low coverage regions, indels and homopolymeric tracts) as soon as high depth Illumina data is available (sequencing ongoing). The released draft genome sequence covers ~92% of the reference sequence. The draft phylogenetic analysis was conducted upon core alignment of unambiguous regions (955 variant positions in a 137 668 bp alignment) retrieved from a rapid alignment (parsnp) of the newly sequenced genome with publicly available genomes (accession numbers are listed in the attached table
Table.zip (632 Bytes)). Data visualization was conducted with Microreact (https://microreact.org/).
- Bunge EM, Hoet B, Chen L, Lienert F, Weidenthaler H, Baer LR, et al. (2022) The changing epidemiology of human monkeypox—A potential threat? A systematic review. PLoS Negl Trop Dis 16(2): e0010141. doi: 10.1371/journal.pntd.0010141
- von Magnus P, Andersen EA, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Path Microbiol Scand. 1959; 46:159.
- Breman JG, Kalisa R, Steniowski MV, Zanotto E, Gromyko AI, Arita I. Human monkeypox, 1970–79. Bull World Health Organ. 1980; 58(2):165–182.
- Mauldin MR, McCollum AM, Nakazawa YJ, Mandra A, Whitehouse ER, et al. Exportation of Monkeypox Virus From the African Continent. J Infect Dis. 2022 Apr 19;225(8):1367-1376. doi: 10.1093/infdis/jiaa559.
- Yinka-Ogunleye A, Aruna O, Dalhat M, et al. Outbreak of human monkeypox in Nigeria in 2017–18: a clinical and epidemiological report. Lancet Infect Dis 2019; 19:872–9. doi: 10.1016/S1473-3099(19)30294-4