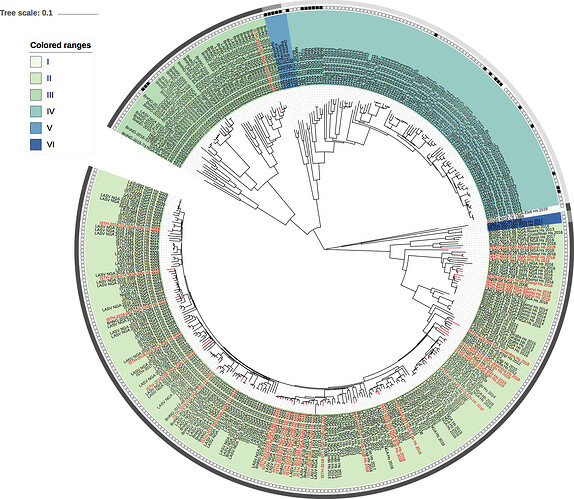
The 35 2018 genomes have now been put together with unpublished Lassa virus sequences (partial and complete) from previous years, that are under analysis by BNITM, and with the 64 2015-2016 genomes recently posted by colleagues at http://virological.org/t/new-lassa-virus-genomes-from-nigeria-2015-2016/191.
11 duplicate genomes from the same samples have been removed. Maximum likelihood phylogenetic analyses of the coding genes in both the L and S segments confirm previous conclusions about the independent spill-over from the rodent reservoir.
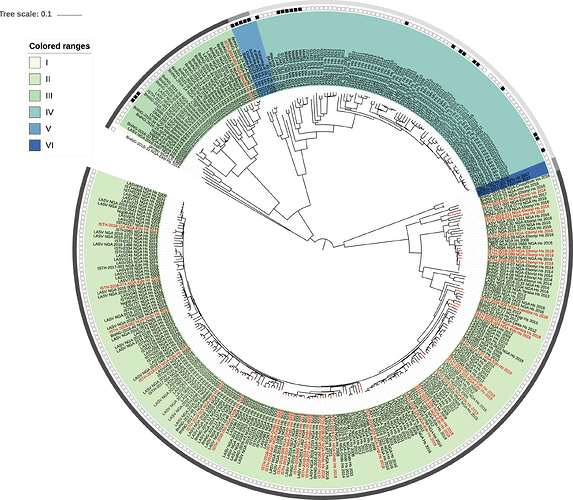
Figure 1: Lassa virus S-segment phylogeny. The circular tree includes all LASV lineages (I-VI), but the clade for lineage IV and V is collapsed for clarity. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. The outer circle represents geographic origin: dark grey = Nigeria, lighter grey = other (Togo, Benin & Mali). Newly sequenced viruses are depicted in red.
Figure 2: Lassa virus L-segment phylogeny. The circular tree includes all LASV lineages (I-VI), but the clade for lineage IV and V is collapsed for clarity. The squares at the tips represent host: empty square = human virus; black square = rodent virus; no square = laboratory virus. The outer circle represents geographic origin: dark grey = Nigeria, lighter grey = other (Togo, Benin & Mali). Newly sequenced viruses are depicted in red.
Acknowledgements.
The Irrua specialist Teaching Hospital (ISTH), Institute for Lassa Fever Research and Control (ILFRC), Irrua, Edo State, Nigeria
The Bernhard-Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany,
Public Health England (PHE),
African Center of Excellence for Genomics of Infectious Disease (ACEGID Redeemer’s University, Ede, Nigeria),
Broad Institute of MIT and Harvard (Cambridge, MA, USA), Harvard University (Cambridge, MA, USA)