There aren’t any Zika virus solved protein structures. I’m proposing to use homology modelling to create some stand-ins until such times as the real thing can be produced. The templates could be various flaviviruses - at the moment I think that dengue is the best bet for most proteins. Around two-thirds of the proteins could be modelled. The models could be used to examine the functional/structual implications of any clade-specific substitutions.
I’d like to consult the community on which Zika genome to model. I’ve been following the conversations on the site about unreliability of some deposited sequences. Normally, for something like this, I’d choose a reference genome for the basic homology model and then locally model substutions in newer genomes. However, I gather that the reference genome MR766 is probably not the best starting point.
Any suggestions much appreciated.
Just to let you know that structural homology modelling already underway here with Thomas Bowden. The more eyes the better though. The 2015/2016 S American strains are all similar, so take consensus or pick a couple of different ones to see if results vary.
Oli
Here’s Zika Haiti capsid modelled on Dengue 1R6R.
WNV might be a better template for capsid. Will try that tomorrow.
…
and here’s the capsid also modelled on WNV 1SFK. This is energetically a much better model.
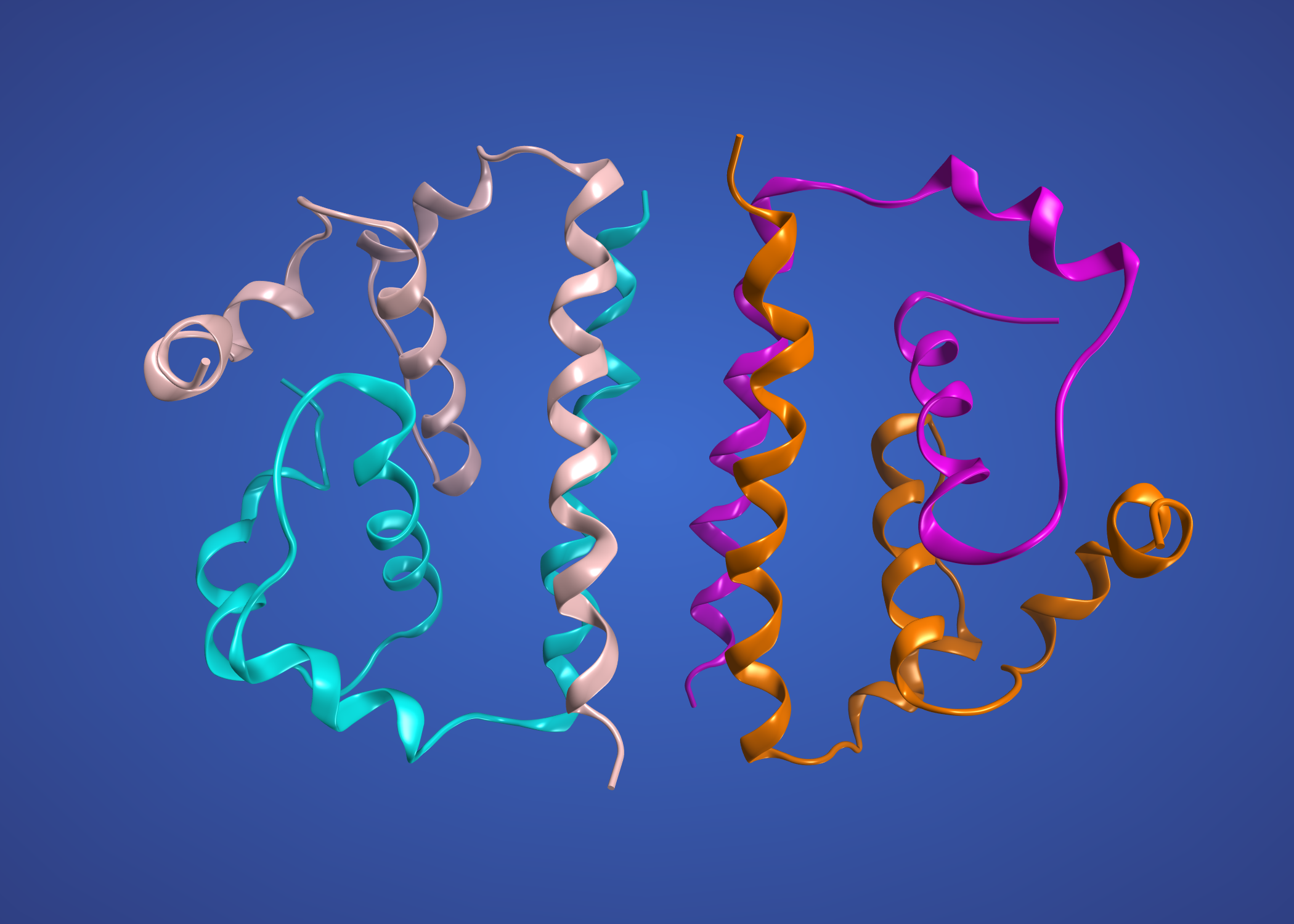
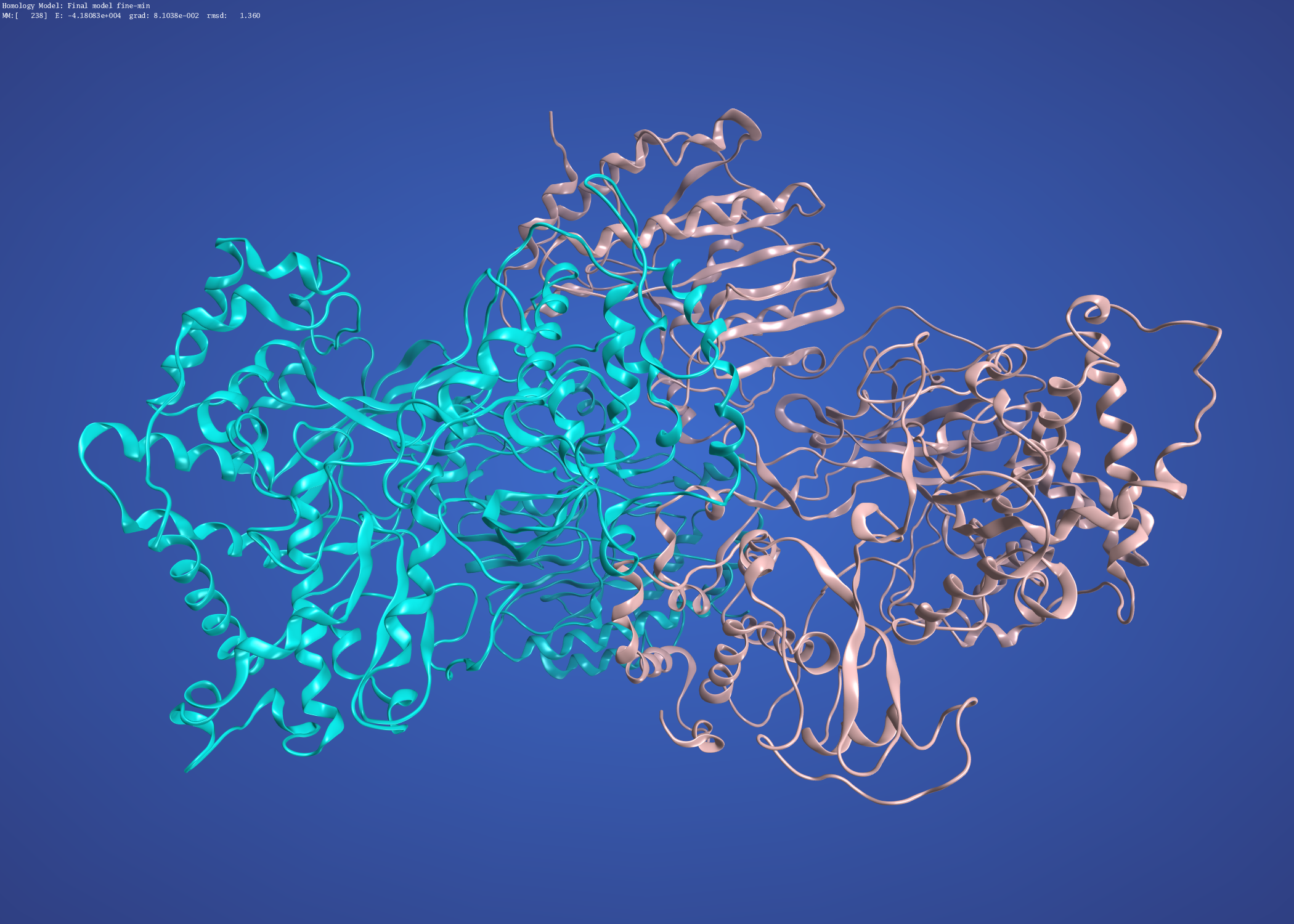
and the precursor membrane protein / envelope protein heterodimer modelled on Dengue 3C5X
This one is NS1 modelled on 4O6C, WNV again.
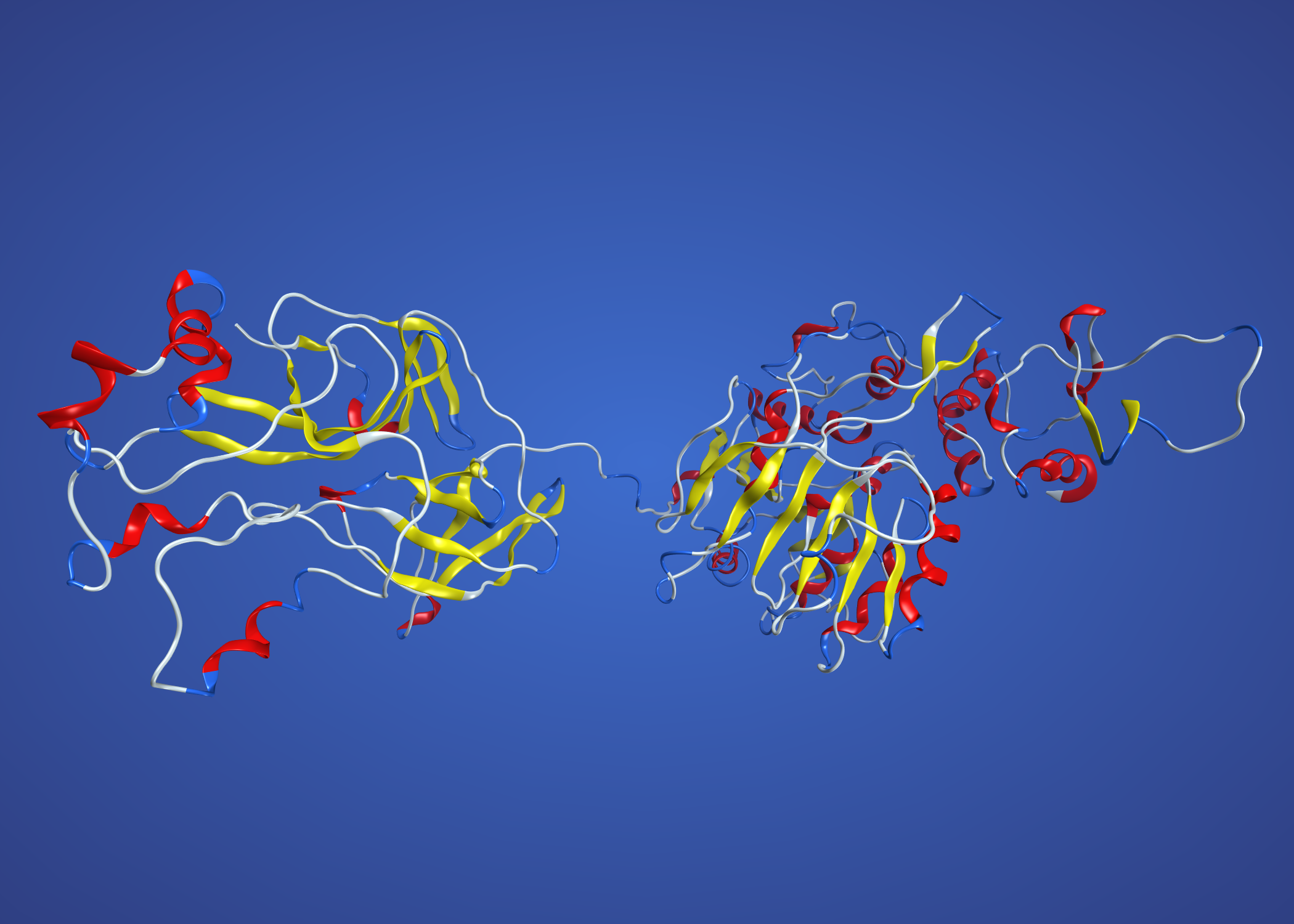
NS3 modelled on 2WV9 Murray Valley virus. This one is single chain, so colouring by secondary structure. The unstructured bit in the middle is an artefact, as the Zika protein is longer (insertion?) in this region and there is no corresponding part of 2WV9 to model on.
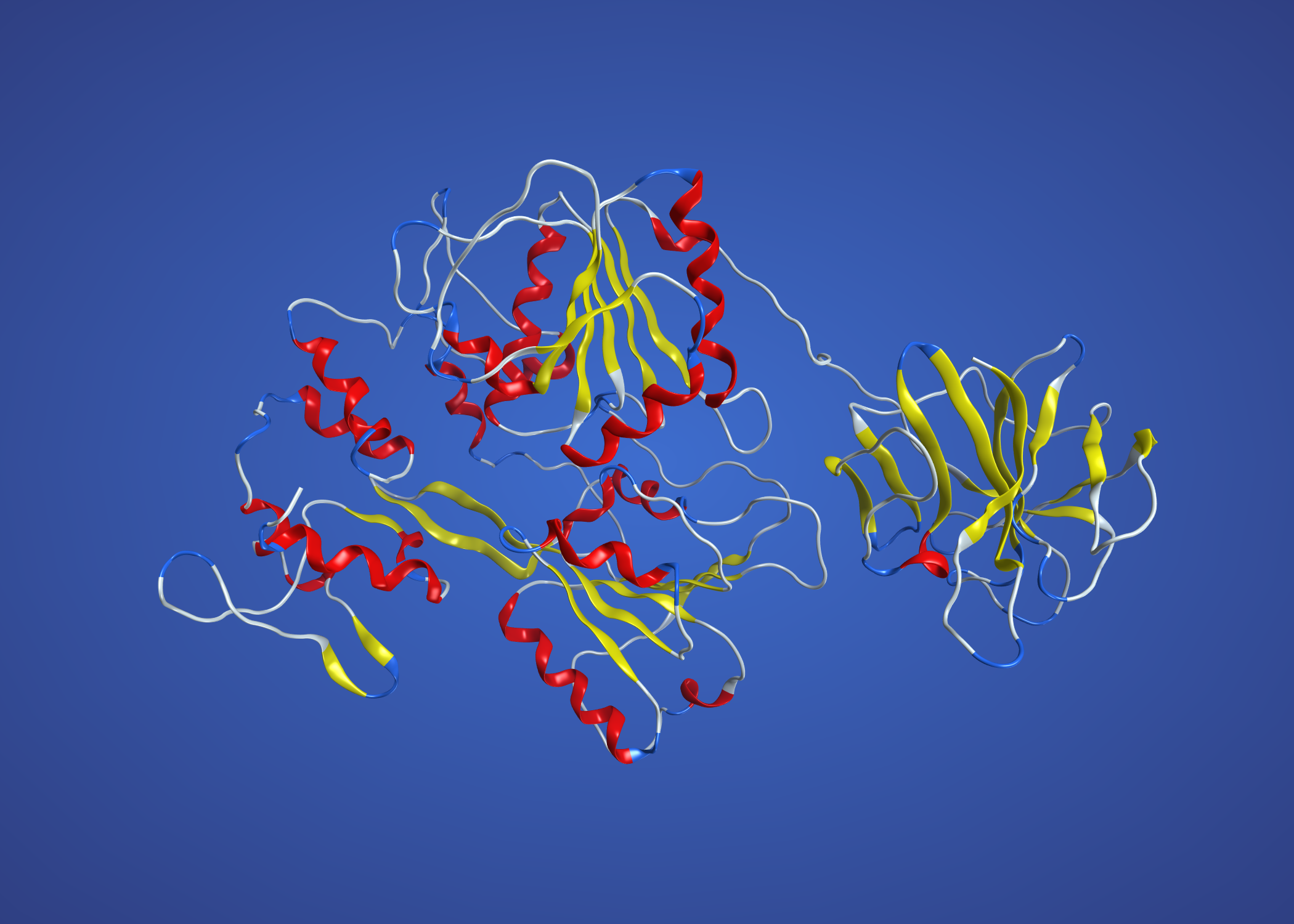
This uses NS3 from dengue, 2WZQ, and is definitely better than the above
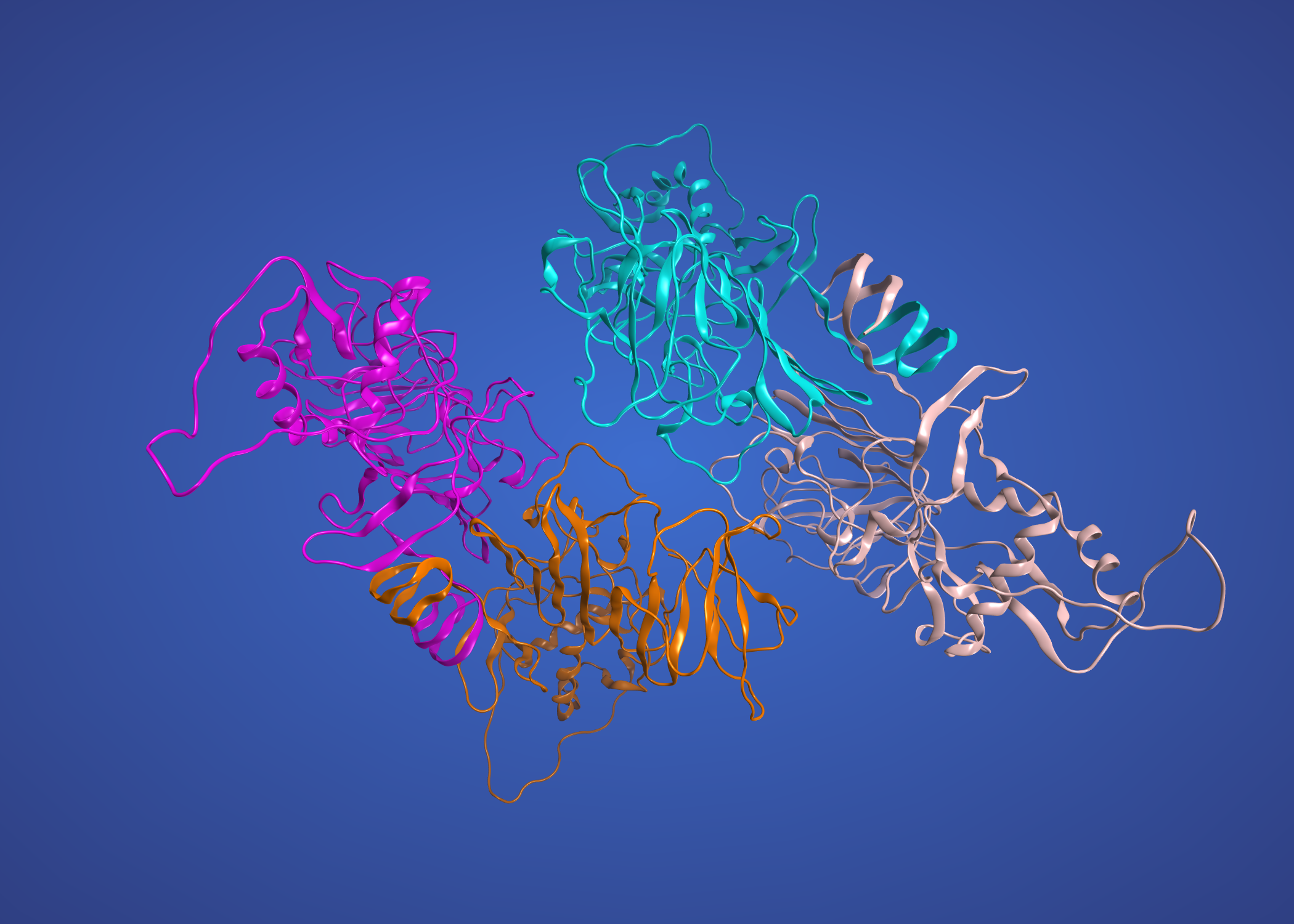
and finally NS5 on 4K6M (Japanese encephalitis)