This is an analysis of Ebola virus genome sequences being produced in the field by teams in Sierra Leone and Guinea over the last few months as the epidemic continues in these countries. A total of 145 sequences from Sierra Leone have been produced a consortium from the Wellcome Trust Sanger Centre, the University of Cambridge and Public Health England. A total of 51 sequences from Guinea have been produced by the EMLab/EVIDENT consortium in collaboration with University of Birmingham.
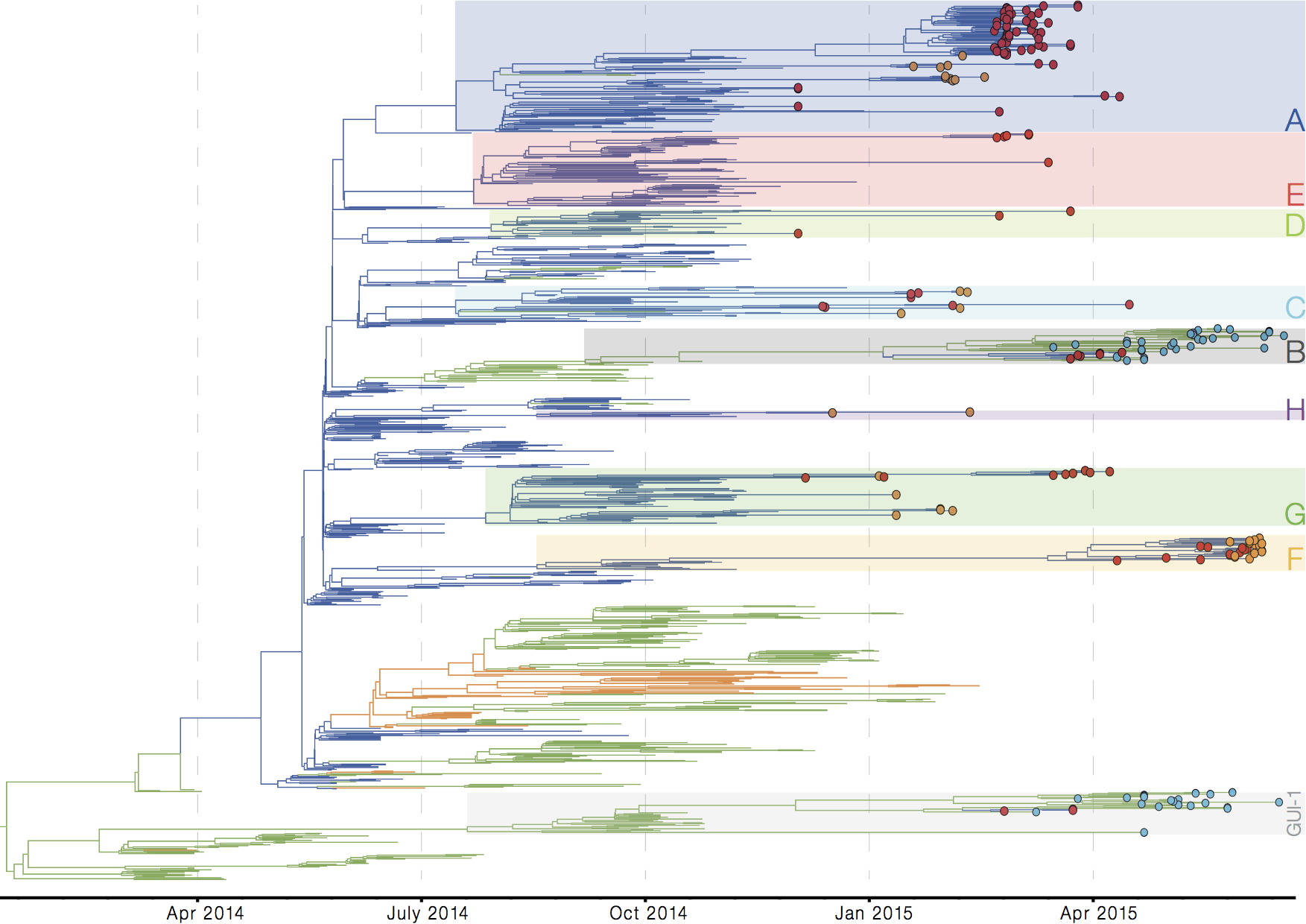
Figure 1 | Time-resolved phylogenetic tree inferred from an alignment of 985 EBOV genomes. Tips coloured in red are from the previous release of WTSI/UoC/PHE genomes, while those in orange are the novel WTSI/UoC/PHE sequences from Sierra Leone. Tips coloured in blue are the novel EMLab-RT sequences from Guinea. Lineages containing the newly-sequenced isolates are highlighted with coloured boxes and labelled A-H for lineages derived from the SL3 lineage, or GUI-1 for the divergent Guinean lineage. Branches are coloured by inferred geographical location: blue for Sierra Leone, green for Guinea, and orange for Liberia.
http://virological.org/uploads/default/original/1X/26d41c9da44bca59e7b2f4a236660e51c7f13840.pdf
Methods. All available West African EBOV genomes from GenBank (1-8) were aligned with the novel genome set. Genomes were partitioned into coding regions and intergenic regions, with the coding region further partitioned into the first, second, and third coding positions. To each partition, an HKY+Γ substitution was employed. An uncorrelated lognormal relaxed clock and a Bayesian SkyGrid tree prior were used. A maximum clade credibility tree was constructed from the posterior set of 9,000 trees drawn from an MCMC chain length of 50M states.
References
- Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, et al. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369-1372.
- Tong YG, Shi WF, Di L, Qian J, Liang L, Bo XC, Liu J, Ren HG, Fan H, Ni M, Sun Y, Jin Y, Teng Y, Li Z, Kargbo D, Dafae F, Kanu A, Chen CC, Lan ZH, Jiang H, Luo Y, Lu HJ, Zhang XG, Yang F, Hu Y, Cao YX, Deng YQ, Su HX, Sun Y, Liu WS, Wang Z, Wang CY, Bu ZY, Guo ZD, Zhang LB, Nie WM, Bai CQ, Sun CH, An XP, Xu PS, Zhang XL, Huang Y, Mi ZQ, Yu D, Yao HW, Feng Y, Xia ZP, Zheng XX, Yang ST, Lu B, et al. 2015. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature doi:10.1038/nature14490.
- Carroll MW, Matthews DA, Hiscox JA, Elmore MJ, Pollakis G, Rambaut A, Hewson R, Garcia-Dorival I, Bore JA, Koundouno R, Abdellati S, Afrough B, Aiyepada J, Akhilomen P, Asogun D, Atkinson B, Badusche M, Bah A, Bate S, Baumann J, Becker D, Becker-Ziaja B, Bocquin A, Borremans B, Bosworth A, Boettcher JP, Cannas A, Carletti F, Castilletti C, Clark S, Colavita F, Diederich S, Donatus A, Duraffour S, Ehichioya D, Ellerbrok H, Fenandez-Garcia MD, Fizet A, Fleischmann E, Gryseels S, Hermelink A, Hinzmann J, Hopf-Guevara U, Ighodalo Y, Jameson L, Kelterbaum A, Kis Z, Kloth S, Kohl C, Korva M, et al. 2015. Temporal and spatial analysis of the 2014-2015 Ebola virus outbreak in West Africa. Nature doi:10.1038/nature14594.
- Park DJ, Dudas G, Wohl S, Goba A, Whitmer SL, Andersen KG, Sealfon RS, Ladner JT, Kugelman JR, Matranga CB, Winnicki SM, Qu J, Gire SK, Gladden-Young A, Jalloh S, Nosamiefan D, Yozwiak NL, Moses LM, Jiang PP, Lin AE, Schaffner SF, Bird B, Towner J, Mamoh M, Gbakie M, Kanneh L, Kargbo D, Massally JL, Kamara FK, Konuwa E, Sellu J, Jalloh AA, Mustapha I, Foday M, Yillah M, Erickson BR, Sealy T, Blau D, Paddock C, Brault A, Amman B, Basile J, Bearden S, Belser J, Bergeron E, Campbell S, Chakrabarti A, Dodd K, Flint M, Gibbons A, et al. 2015. Ebola Virus Epidemiology, Transmission, and Evolution during Seven Months in Sierra Leone. Cell 161:1516-1526.
- Simon-Loriere E, Faye O, Faye O, Koivogui L, Magassouba N, Keita S, Thiberge JM, Diancourt L, Bouchier C, Vandenbogaert M, Caro V, Fall G, Buchmann JP, Matranga CB, Sabeti PC, Manuguerra JC, Holmes EC, Sall AA. 2015. Distinct lineages of Ebola virus in Guinea during the 2014 West African epidemic. Nature doi:10.1038/nature14612.
- Kugelman JR, Wiley MR, Mate S, Ladner JT, Beitzel B, Fakoli L, Taweh F, Prieto K, Diclaro JW, Minogue T, Schoepp RJ, Schaecher KE, Pettitt J, Bateman S, Fair J, Kuhn JH, Hensley L, Park DJ, Sabeti PC, Sanchez-Lockhart M, Bolay FK, Palacios G, Diseases USAMRIoI, National Institutes of H, Integrated Research Facility-Frederick Ebola Response T. 2015. Monitoring of Ebola Virus Makona Evolution through Establishment of Advanced Genomic Capability in Liberia. Emerg Infect Dis 21:1135-1143.
- Bell A, Lewandowski K, Myers R, Wooldridge D, Aarons E, Simpson A, Vipond R, Jacobs M, Gharbia S, Zambon M. 2015. Genome sequence analysis of Ebola virus in clinical samples from three British healthcare workers, August 2014 to March 2015. Euro Surveill 20.
- Hoenen T, Safronetz D, Groseth A, Wollenberg KR, Koita OA, Diarra B, Fall IS, Haidara FC, Diallo F, Sanogo M, Sarro YS, Kone A, Togo AC, Traore A, Kodio M, Dosseh A, Rosenke K, de Wit E, Feldmann F, Ebihara H, Munster VJ, Zoon KC, Feldmann H, Sow S. 2015. Virology. Mutation rate and genotype variation of Ebola virus from Mali case sequences. Science 348:117-119.
This is interesting. It appears to me that there’s more diversity among the lineages from Sierra Leone than from the other two countries. Is that correct? If so, is that simply a reflection of our sampling bias or is it possible that we have many more transmission chains ongoing in Sierra Leone, whereas transmission chains are more localized in Guinea? (and more so in Liberia).
Also, a lot of endemic circulation going on.
The pattern in Sierra Leone looks like what we would expect as a large epidemic declines - the gradual extinction of diversity that accumulated during the exponential growth. But it didn’t decline to zero so the lineages that persisted then start to accumulate diversity again. Guinea has fewer of these lineages as it was never the peaking epidemic that the SLE and LBR had.
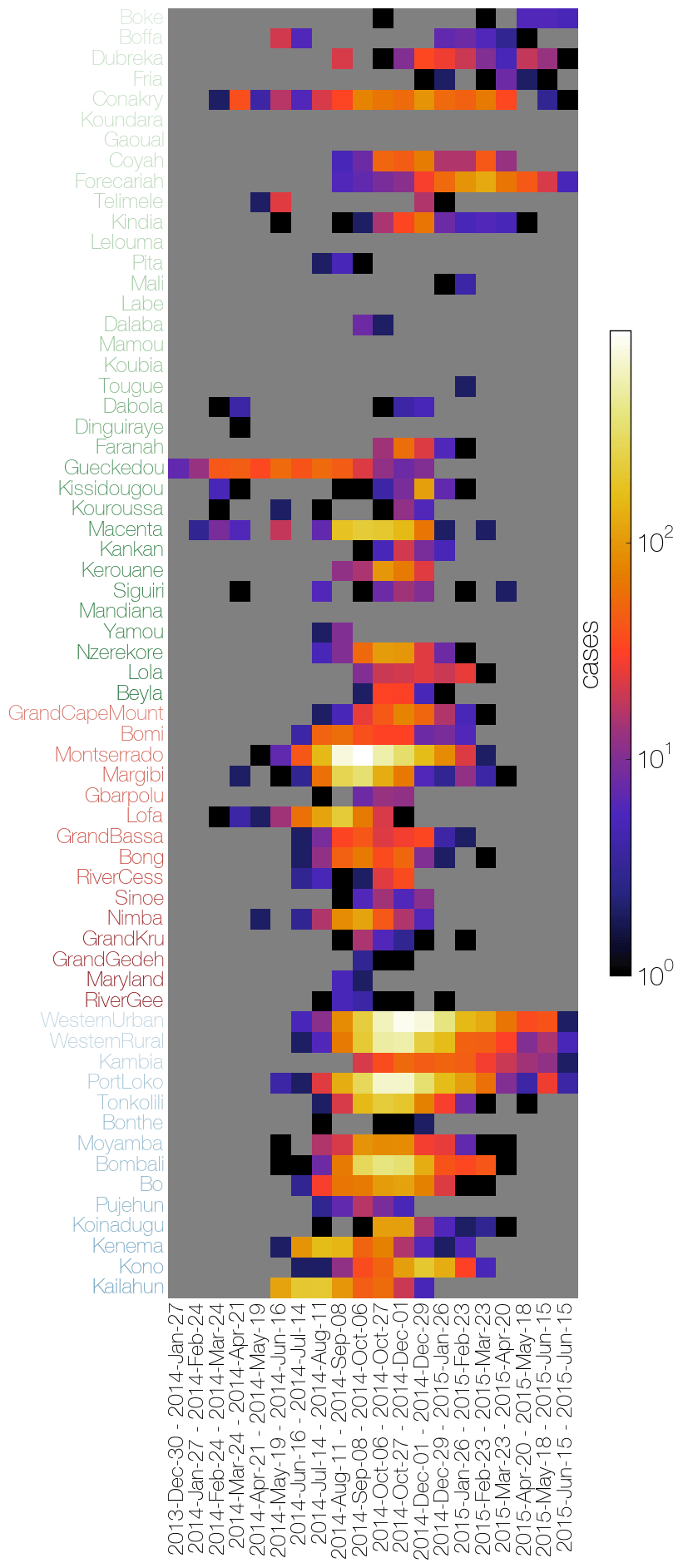
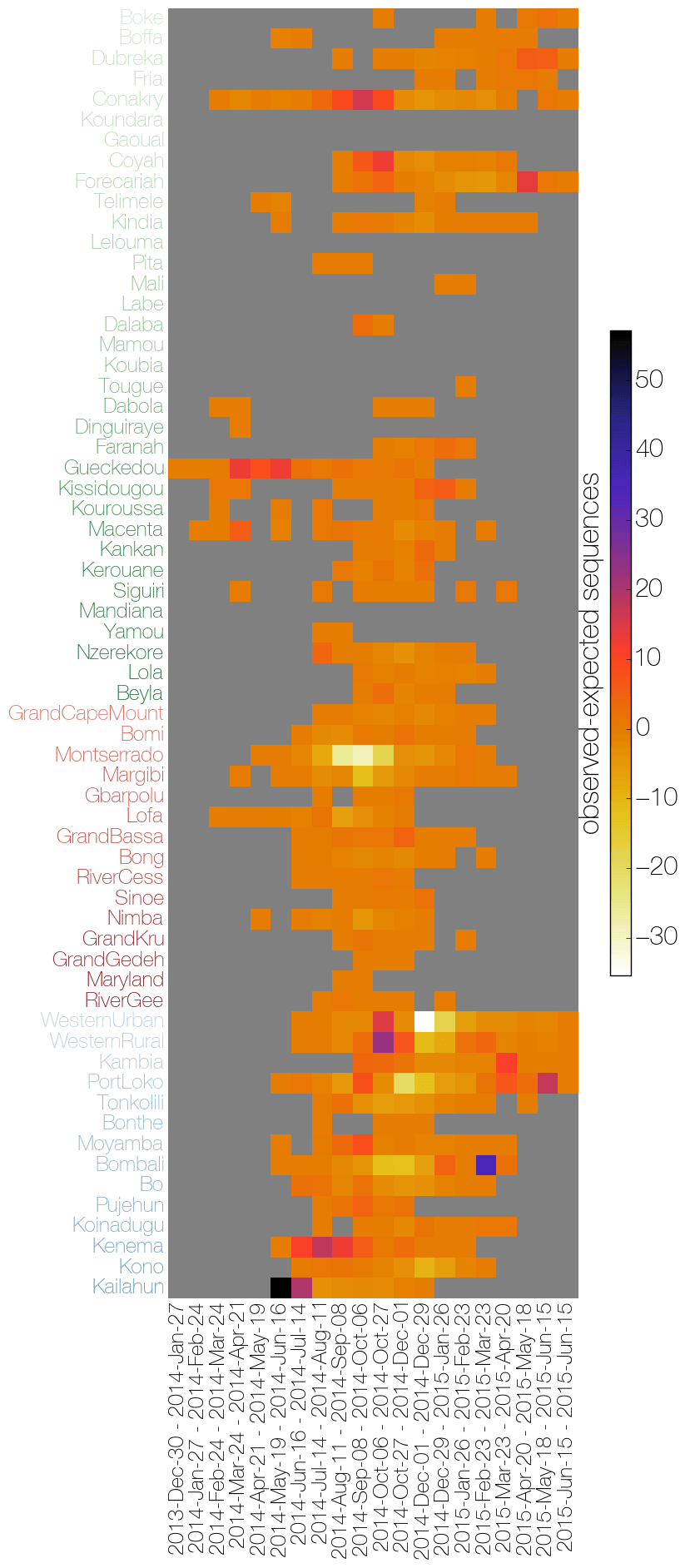
Heatmaps of the number of cases reported by the WHO (left) and the viral genome sampling by all agencies (right). The right hand plot shows the difference in the actual numbers of sequences by location and month to the expected number if distributed proportionally to the case counts.
Plots by @evogytis